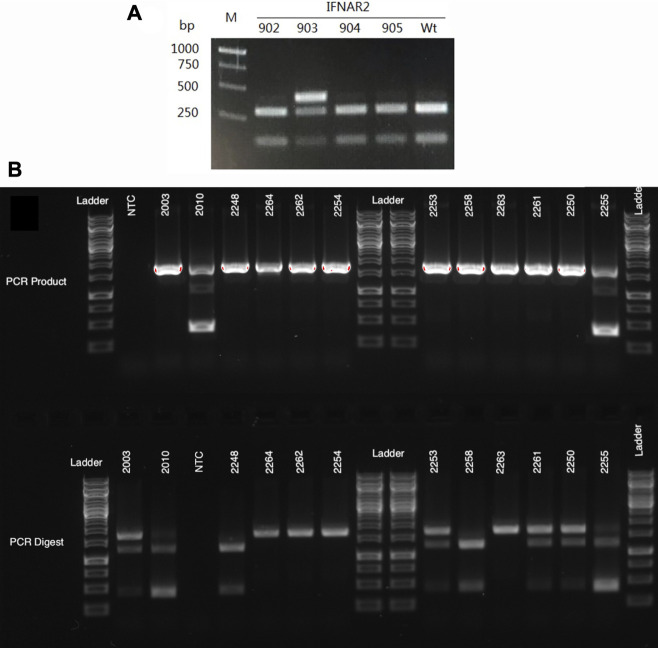
FIGURE 2.
Genotyping gels for IFNAR2 knockout sheep. (A) An example of the initial PCR-RFLP typing results for offspring of 7IFN2, an IFNAR2 −/− ram. PCR amplification was done with primers created for screening the knockout cell colonies that amplify a 418 bp segment of the gene. Since the sire was a biallelic knockout and the dams were WT Romneys, the lambs were all expected to have one WT and one knockout allele. However, three of the four lambs (IFN902, 904 and 905) appeared to only have the WT allele, which is cut by the restriction enzyme NcoI. In contrast, lamb IFN903 had both a WT and an uncut allele with the previously identified 5 bp deletion. These results suggested that the sire carried a second IFNAR2 knockout allele with a large deletion. (B) The top gel shows PCR amplification of genomic DNA with primers that amplify a 1,706 bp segment of the IFNAR2 gene. Two of the sheep on this gel, IFN2010 and IFN2255, have a large 1,282 bp deletion, resulting in a much smaller 424 bp amplicon. The bottom gel shows the PCR products following digestion with NcoI. The WT allele, which is cut by NcoI, appears as two bands of 1,259 and 447 bp. The knockout allele with a small 6 bp deletion remains uncut and is seen as a 1,701 bp band.