Figure 2. A subset of tumors has cell-associated bacteria.
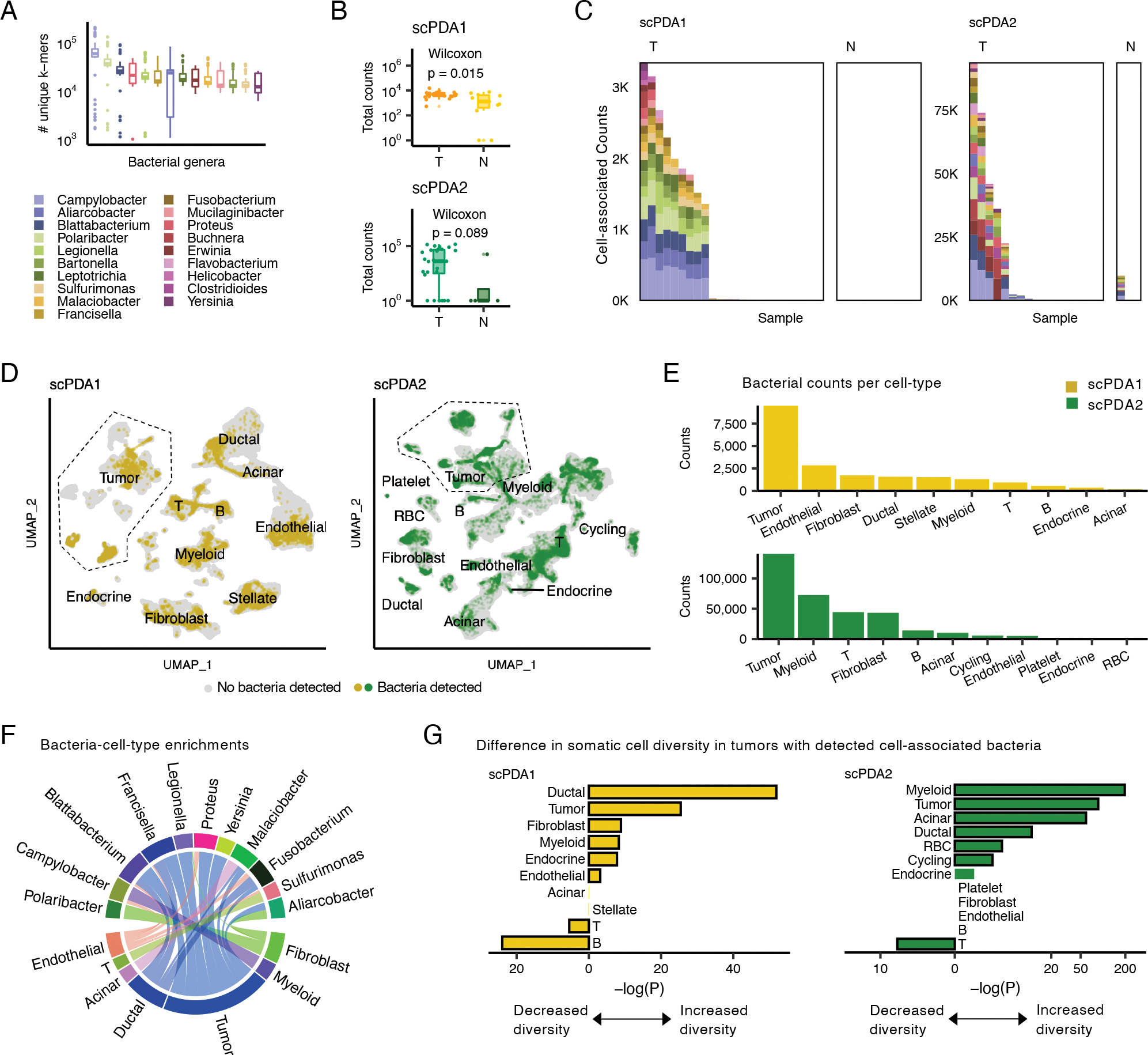
(A) Boxplots showing the number of unique k-mers assigned to each genus detected in scPDA1-2. Boxplots show median (line), 25th and 75th percentiles (box) and 1.5xIQR (whiskers); points represent outliers. (B) Boxplots (top: scPDA1, bottom: scPDA2) comparing the total number of bacterial counts per sample in T (tumor) vs. N (normal) samples. Boxplots as in (A). (C) Profiles of cell-associated bacterial counts in scPDA1 and 2. Stacked bar plots showing the number of counts and genus composition for each sample. K, thousand; T, tumor samples; N, normal samples. (D) Uniform manifold approximation and project (UMAP) plots of host somatic cells for scPDA1 (n=57,530 cells) and scPDA2 (n=59,473 cells). Clusters are labeled by cell type. Cells are colored yellow (scPDA1) or green (scPDA2) if they share a barcode with bacteria. (E) Bar plots of the number of bacterial counts associated with each cell type in scPDA1-2. (F) Circos-plot of significant (p<5e-3) bacteria-cell-type enrichments identified at the single-barcode level by Wilcoxon testing and shared in scPDA1-2. Ribbon width correlates with enrichment strength. (G) Bar plots showing the adjusted Wilcoxon p-value comparing the transcriptional diversity of bacteria-associated cells to unassociated cells for each cell type. See also Figure S2.
