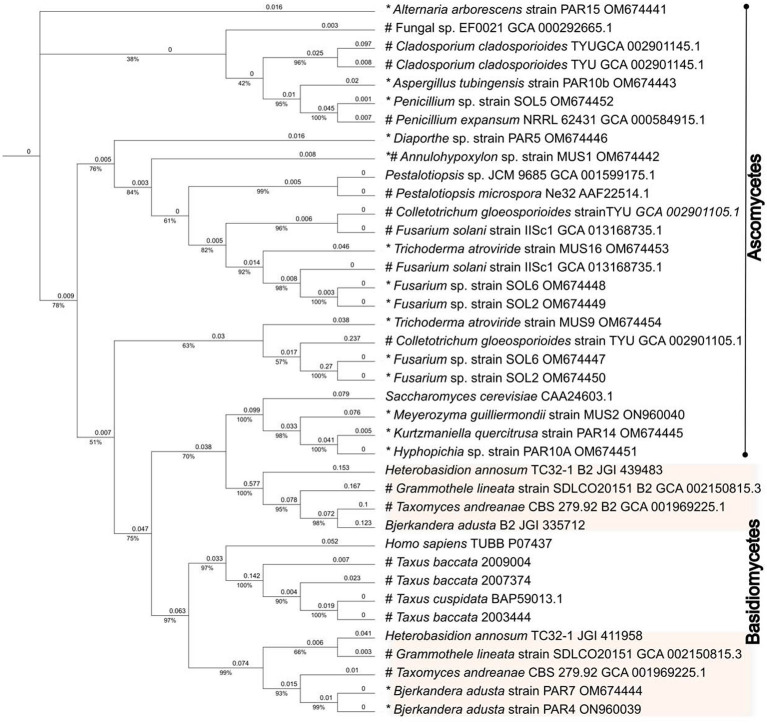
Figure 4.
Evolutionary analysis of the β-tubulins proteins. The evolutionary history was inferred by using the IQ-TREE web server (Trifinopoulos et al., 2016) and edited using the Interactive Tree Of Life (iTOL) online tool (Letunic and Bork, 2021). The analysis was done using the default parameters. The analysis only included the 428 amino acids that have been implicated in the paclitaxel-β-tubulin interaction. In addition, GenBank protein sequences from fungal endophytes isolated from Taxus spp. or from fungi deemed able to synthesize paclitaxel were included in the analysis. For comparison, the human and Taxus β-tubulins protein sequences were also included. The phylogenetic tree generated formed branches that clearly allocated the β-tubulins along the Ascomycetes and Basidiomycetes divisions. The β-tubulins from Taxus spp. and the most highly expressed human β-tubulin (P07437) clustered together in a separate branch with the Basidiomycete β-tubulins. The Taxomyces andreanae β1-tubulin also clustered in this group, and the β2 tubulin clustered with the rest of the β2 tubulin from Basidiomycete fungi in a separate clade. This would suggest that T. andreanae CBS 279.92 belongs in the phylum Basidiomycota, rather than Ascomycota as originally proposed. An asterisk (*) indicates the fungi isolated by Gauchan et al. (2021), which were used in this study, and the pound sign (#) indicates plant/fungus that can synthesize paclitaxel. JGI numbers refer to protein sequence number available from the Joint Genome Institute (https://mycocosm.jgi.doe.gov/mycocosm/home).