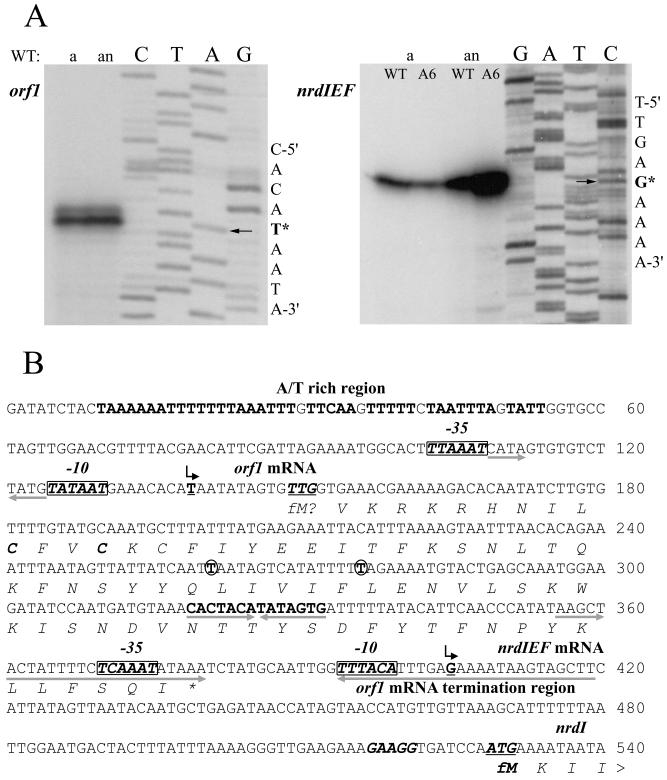
FIG. 4.
(A) Primer extension analysis of orf1 and nrdIEF genes. Total RNA was isolated from aerobic (a) and anaerobic (an) cultures of RN4220 (WT) and the MMA6 mutant (A6). Primer extension was carried out with the primers orf1-rev and nrdI-rev (see Materials and Methods), and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions using the same primers. The arrows point to the nucleotides (labeled with an asterisk) of the orf1 and nrdIEF transcription start points. (B) Nucleotide sequence of the orf1 and nrdIEF promoter regions. Transcription start points are shown by bent arrows above the underlined boldface T nucleotide (for orf1) and G nucleotide (for nrdIEF) start sites. The nrdI ATG translational start codon (underlined) and its ribosomal binding site are shown in boldface italic letters; a putative orf1 translational start codon, TTG (underlined), and two in-frame cysteines are also shown in boldface italics. Putative −10 and −35 sequences are shown as boxed boldface italic letters. The pairs of opposing arrows placed beneath nucleotide sequences show three inverted-repeat sequences. The circled nucleotides are those that are changed in the orf1 of some S. aureus strains (see the text); another feature is a long A/T rich region (boldface roman letters) upstream of orf1.