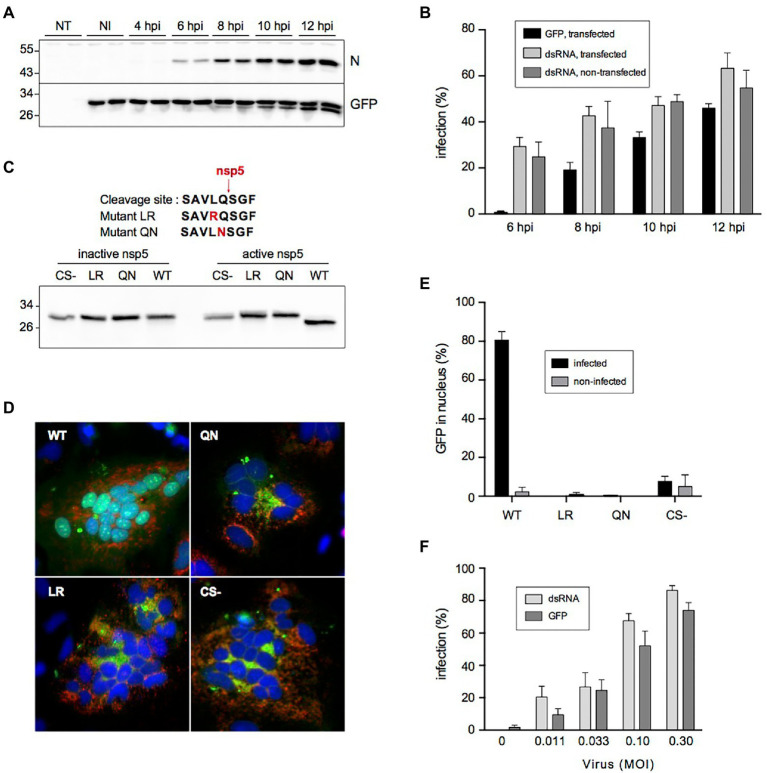
Figure 3.
Cleavage of the fluorescent reporter in SARS-CoV-2-infected cells. (A) Kinetics of cleavage analyzed by immunoblotting. NT: non-transfected, NI: non-infected, N: nucleocapsid, GFP: fluorescent reporter detected with an antibody to GFP. (B) Kinetics of infection quantified by nuclear translocation (GFP) in cells transfected with the fluorescent reporter and by dsRNA immunostaining in transfected or non-transfected cells (mean ± SD of 6 replicates). (C) Impact of mutating Leu and Gln residues on the cleavage by SARS-CoV-2 nsp5. The amino acid sequences of the mutant cleavage sites are shown on top. HeLa cells were co-transfected with indicated constructs and an active or inactive version of nsp5 and analyzed by immunoblotting with an antibody to GFP. CS- is a construct lacking the whole cleavage site peptide. (D) Impact of cleavage site mutation on nuclear transfer of the reporter. HeLa cells were co-transfected with indicated constructs (green) and a plasmid expressing human ACE2 and infected with SARS-CoV-2. Cells were fixed and processed for immunofluorescent detection of dsRNA (red) and DAPI staining (blue) at 16 hpi. (E) Impact of cleavage site mutation on nuclear transfer of the reporter in Vero cells. Vero-81-derived clone 6 cells were transfected with indicated constructs and infected with SARS-CoV-2. Infected cells were defined by dsRNA immunostaining and the nuclear localization of GFP in infected and non-infected cells was quantified (mean ± SEM of 4 independent experiments). (F) Nuclear GFP staining as a function of virus input. The infection was measured by counting cells GFP staining (GFP) or by immunostaining to dsRNA (mean ± SD of 8 replicates).