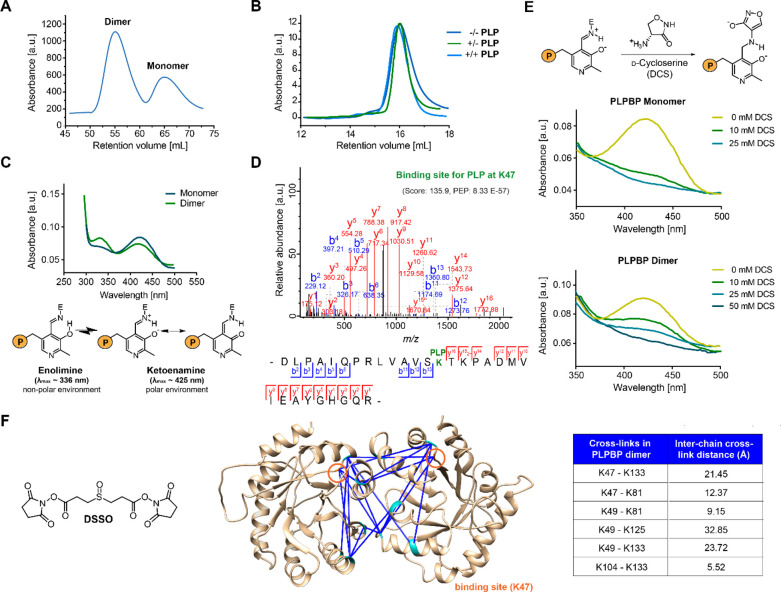
Figure 1.
Properties of recombinant human PLPBP. (A) The main oligomeric state of tag-free PLPBP is dimeric, with a small fraction of monomer according to size-exclusion chromatography (SEC; preparative scale). (B) Preincubation of monomeric PLPBP with either excess PLP and separation in buffer without PLP (+/–) or in buffer containing PLP (+/+) or preincubation and separation in the absence of PLP (−/–; analytical scale; retention volume of dimer: approximately 13.82 mL). (C) UV/vis spectra of PLPBP monomer and dimer display characteristic absorbance peaks corresponding to the internal aldimine. Peak intensities resulting from the enolimine (λmax ≈ 336 nm) and ketoenamine (λmax ≈ 425 nm) forms of the internal aldimine were different for both monomers and dimers at equal concentrations (P = OPO32–).21 (D) Tandem MS (MS/MS) spectra identifying the PLP-binding site of PLPBP (K47) with corresponding score and posterior error probability (PEP; false discovery rate, FDR < 0.01). (E) Incubation with 10 mM of the PLP-binding antibiotic d-cycloserine (DCS) leads to almost complete displacement of PLP from monomeric PLPBP, whereas 50 mM DCS was required for the PLPBP dimer (P = OPO32–).22 (F) Analysis of PLPBP by chemical cross-linking combined with mass spectrometry (XL-MS) utilizing the DSSO cross-linker (left). Cross-links solely occurring in the dimer peak after treatment (right, two independent samples (50- and 100-fold excess DSSO) with two fragmentation strategies each, FDR < 0.01) served as restraints for molecular docking of two PLPBP monomers (derived from SWISS-MODEL23 using the yeast ortholog as template, PDB 1B54(13)) using HADDOCK24 to generate a dimer model (center). Median Cα–Cα cross-link distances are all within the maximum distance span of DSSO (35 Å). Lysine residues involved in cross-linking contacts are highlighted cyan, and the PLP binding site is colored orange.