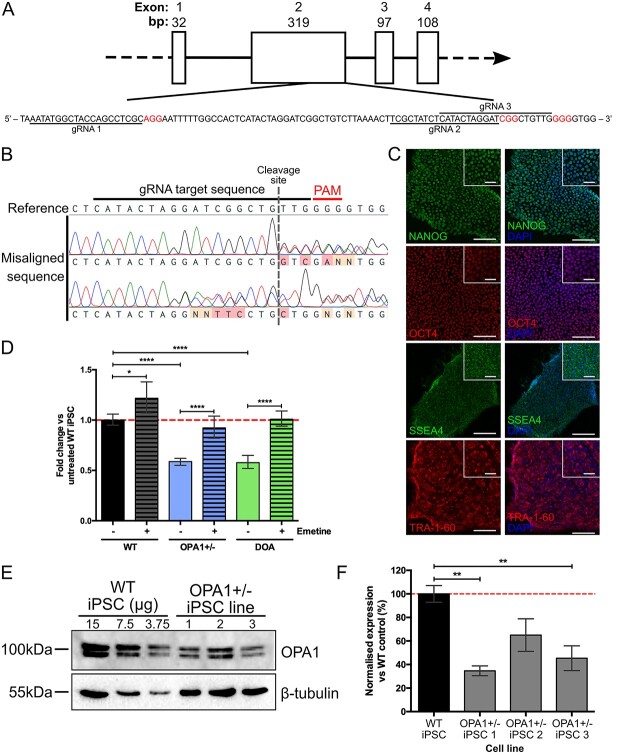
Figure 1.
Generation of isogenic OPA1 heterozygous knockout iPSC. (A) Schematic of OPA1 exons 1–4 and expanded exon 2 sequence showing location of gRNA target sequences (underlined) and protospacer adjacent motif sites (red). (B) Sequence alignment of isolated iPSCs clones to the WT OPA1 sequence enables detection of CRISPR/Cas9 induced mutations (red/orange mis-aligned bases) surrounding the CRISPR/Cas9 gRNA cut site. (C) Isogenic OPA1+/− iPSCs demonstrate nuclear localization of ESC markers NANOG and OCT4 with membrane/cytoplasmic localization of SSEA4 and TRA-1-60. Scale bars = 100 and 40 μm for inset. (D) Expression of total OPA1 in CRISPR/Cas9 edited OPA1+/− and patient-derived DOA iPSCs, with and without emetine, a NMD inhibitor. OPA1 expression was normalized to the geometric mean of GAPDH and ACTIN before normalization to WT expression. n = 4 individual iPSC RNA preparations, ****P < 0.0001. (E) Analysis of OPA1+/− iPSCs OPA1 protein expression via western blot. OPA1 antibody detects short and long isoforms of OPA1 at ~ 100 kDa. β-tubulin reference protein, 55 kDa, used for normalization of OPA1 expression. (F) Three individual OPA1+/− iPSC cell lines show significantly reduced levels of OPA1 protein expression compared to the WT control iPSC, after normalization to reference protein β-tubulin. Bars represent mean ± SEM, n = 3 biological replicates from 3 independent western blots, *P < 0.05, **P < 0.01, ***P < 0.001.