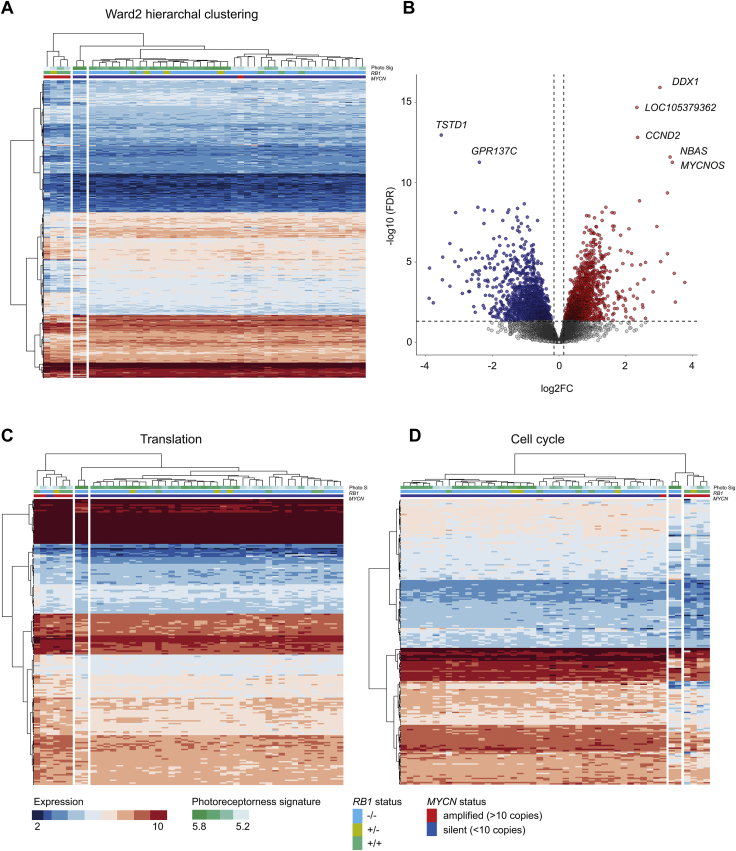
Figure 1.
Differential expression analysis between MYCN-amplified RB1-proficient retinoblastoma tumors and MYCN-silent and RB1-deficient retinoblastoma tumors. MYCN-amplified, RB1-proficient retinoblastoma tumors display significantly different gene expression patterns compared with the MYCN-silent and RB1-deficient samples. A, Heatmap showing hierarchal clustering (WARD2) findings. The relationship between samples based on the 3155 differentially expressed genes in MYCNARB1PRO retinoblastomas is depicted. Rows represent genes. Columns represent samples. Expression colors are adjusted by the level of expression. The MYCN and RB1 status and samples’ photoreceptorness scores are annotated beneath the dendrogram. The MYCNARB1PRO tumors form their own distinct cluster, departing from other retinoblastoma tumors irrelevant of photoreceptorness background. B, Volcano plot showing the differentially expressed genes in MYCNARB1PRO retinoblastomas. Top MYCN-amplified upregulated genes are associated with chromosome 2 amplification. The most significantly downregulated gene in MYCNARB1PRO samples, TSTD1, functions in hydrogen-sulfide metabolism. C, Heatmap showing hierarchical clustering (WARD2) visualizing the expression of 229 ribosomal RNA processing and translation initiation-associated genes significantly upregulated in MYCNARB1PRO samples. The high-level MYCN-amplified tumors cluster together independent of RB1 genotypic background or photoreceptorness levels. D, Heatmap showing hierarchical clustering (WARD2) for the expression of 220 cell cycle-related genes significantly downregulated in MYCNARB1PRO samples. MYCN-amplified samples with RB1-proficient background cluster together. Two RB1-null MYCN-silent samples indicate a closer relationship to MYCNARB1PRO samples and cluster in the adjacent subbranch. Expression values as normalized log2 robust multiarray average: upregulation, log2 fold change (FC) > 0 and adjusted P < 0.05; downregulation, log2 FC < 0 and adjusted P < 0.05. FDR = false dscovery rate.