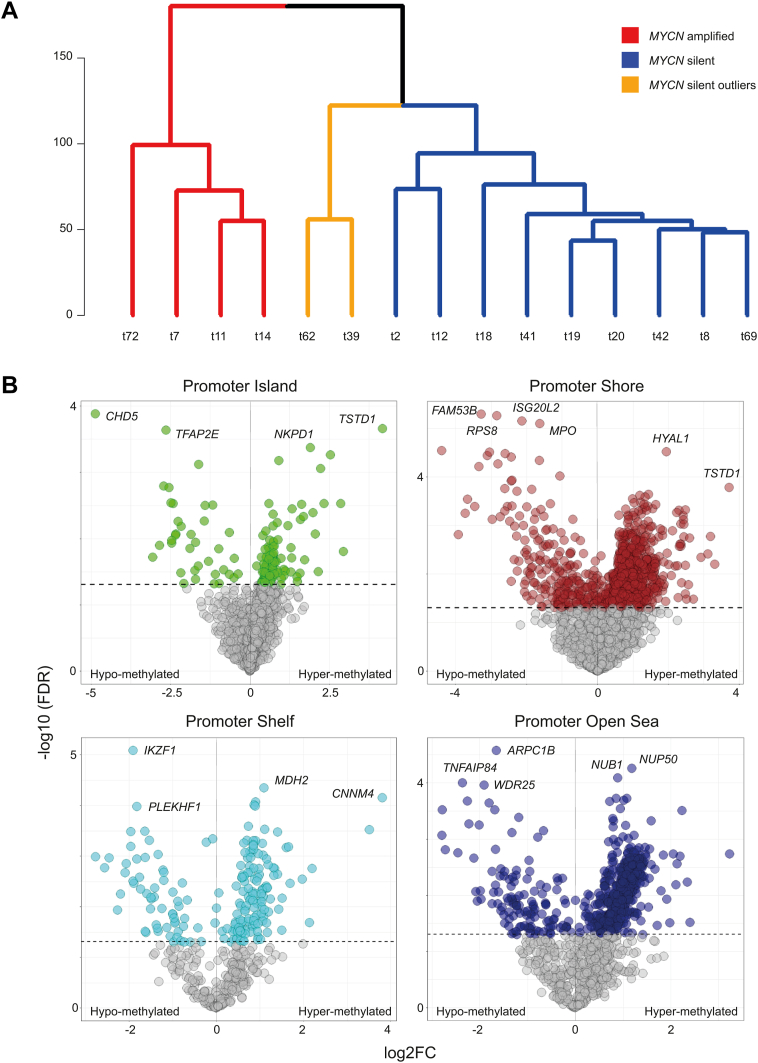
Figure 2.
Differential methylation analysis between a group of 4 high-level MYCN-amplified and a group of 11 MYCN-silent retinoblastomas. MYCN tumors possess distinct methylation patterns compared with non–MYCN-amplified retinoblastomas. A, Dendrogram showing the hierarchical clustering relationships of the retinoblastoma samples. Based on the WARD2 clustering method, MYCN-amplified tumors display distinct methylation patterns compared with MYCN-silent retinoblastomas. B, Volcano plots depicting the differentially methylated genes at various promoter methylation sites between MYCN-amplified and MYCN-silent samples. All sites indicate significantly differentially methylated genes between MYCN-amplified and MYCN-silent samples. Methylated signal is quantile-normalized β values: hypermethylated, log2 fold change (FC) > 0 and adjusted P < 0.05; hypomethylated, log2 FC < 0 and adjusted P < 0.05). FDR = false discovery rate.