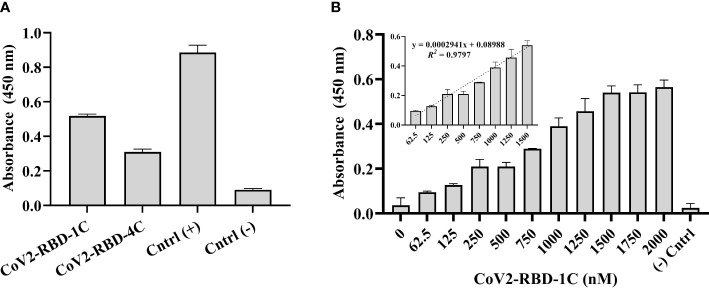
Figure 2.
Screening of biotinylated aptamers binding affinity against SARS-CoV-2 S glycoprotein. (A) Direct ELONA was used to screen two aptamer sequences, CoV2-RBD-1C and CoV2-RBD-4C (1,500 nM), for the detection of S glycoprotein (N = 3). (B) Concentration-dependent analysis of the binding affinity of CoV2-RBD-1C aptamer sequence to SARS-CoV-2 S glycoprotein using direct ELONA (N = 3). The positive control was biotinylated SARS-CoV-2 S monoclonal antibody (30 ng/ml), RBD S glycoprotein coated with no aptamer served as the negative control, and nuclease-free water served as the background control. OD450 was obtained by subtracting the background control from all other wells. The R2 value (B inset) showed that 97% of the absorbance values fitted the regression model. The regression equation for the difference in absorbance value and aptamer concentration (62.5 – 1,500 nM) was y = 0.0002941x + 0.08988, where x is the concentration of the aptamer in nM, and y is the absorbance value (OD450) (B inset). All results are represented in ± SEMs.