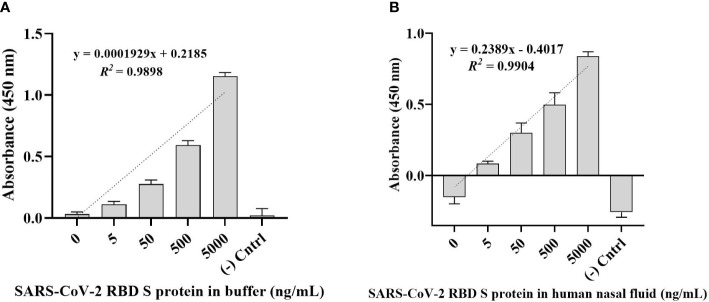
Figure 3.
Detection of SARS-CoV-2 RBD S glycoprotein using CoV2-RBD-1C aptamer-based direct ELONA. (A) SARS-CoV-2 RBD S glycoprotein in a buffer solution (N = 3). Nuclease-free water was used for the background control, while S glycoprotein coated with no aptamer served as the negative control. The regression equation for difference in absorbance value and S glycoprotein concentration (5 - 5,000 ng/ml) in sodium carbonate buffer was y = 0.0001929x + 0.2185 with R2 = 0.98, where x is the concentration of the S glycoprotein in ng/ml, and y is the absorbance value (OD450). (B) SARS-CoV-2 RBD S glycoprotein spiked in a 0.1% human nasal fluid (N = 6). Diluted human nasal fluid was used as the background control, while S glycoprotein in a 0.1% human nasal fluid coated with no aptamer served as the negative control. Absorbance was obtained by subtracting the background control from all other wells. The regression equation for OD450 difference versus S glycoprotein concentration (0 - 5,000 ng/ml) in 0.1% human nasal fluid was y = 0.2389x – 0.4017 with R2 = 0.99. All results are presented in ± SEMs.