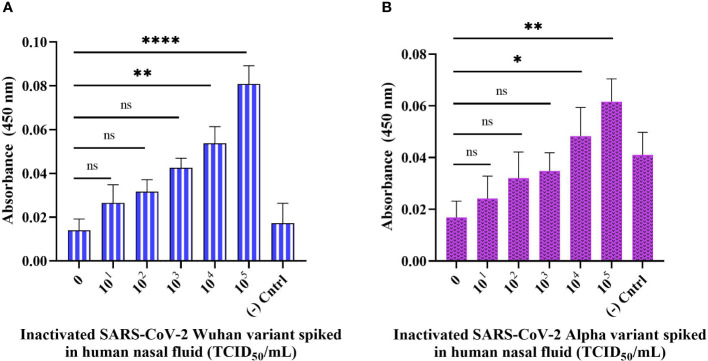
Figure 7.
Detection of inactivated SARS-CoV-2 variants spiked in a 0.1% human nasal fluid using sandwich ELONA. Inactivated SARS-CoV-2 (A) Wuhan variant, and (B) Alpha variant. 0.1% nasal fluid without inactivated viral samples was used as the background control, while each inactivated variant without the aptamers served as the negative control. Absorbance was obtained by subtracting the background control from all other wells where the viral samples were coated. All data are presented in ± SEMs (N = 6), and the statistically significant differences between the viral titer tested was determined: ns – not significant, *p< 0.05, **p< 0.01, ****p< 0.0001. The regression equation for difference in absorbance value and Wuhan variant viral titer (0 to 105 TCID50/mL) in 0.1% human nasal fluid was y = 0.01219x + 0.01106 with R2 = 0.93, while for Alpha variant the regression equation was y = 0.008557x + 0.01491 with R2 = 0.95, where x is the concentration of the inactivated variants in TCID50/mL, and y is the absorbance value (OD450).