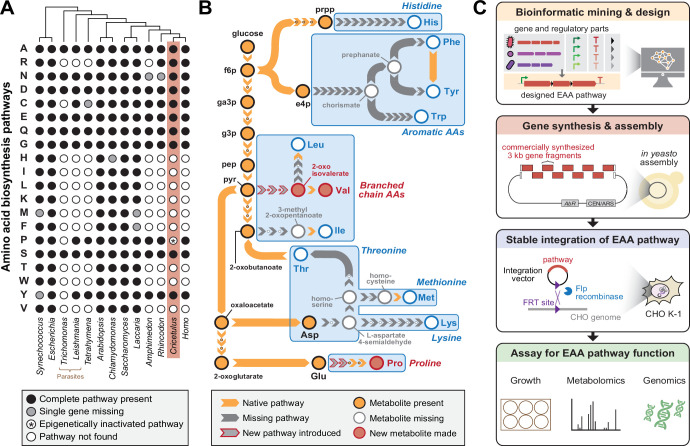
Figure 1. Engineering Essential Amino Acid (EAA) biosynthesis in metazoan cells.
(A) Presence of amino acid biosynthesis pathways across representative diverse organisms on Earth. (B) Schematic of EAA biosynthesis pathway steps that require engineering in mammalian cells to enable complete amino acid prototrophy if imported from Escherichia coli. Proline and Valine pathways shown in this work are highlighted in red. (C) Workflow diagram of a synthetic genomics approach involving pathway design, construction, integration and testing towards mammalian EAA restoration.