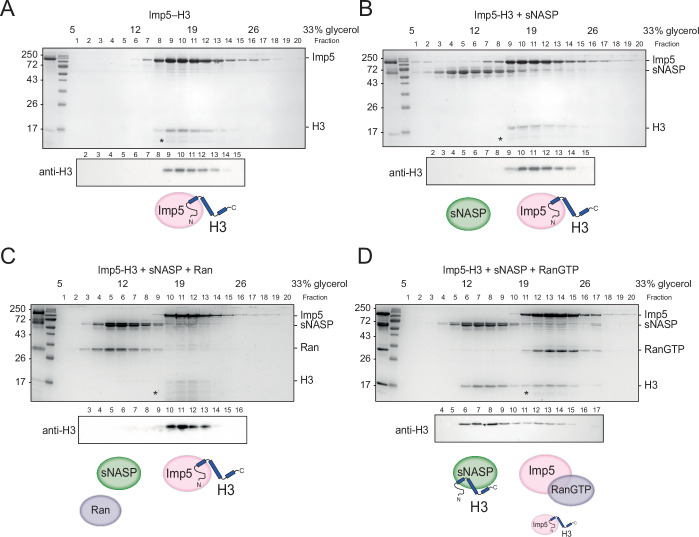
Figure 4. H3 binding by Imp5 and sNASP is mutually exclusive and relies on RanGTP for histone transfer.
(A) Imp5-H3 reconstituted complex was purified by size-exclusion chromatography (SEC) and incubated on ice (control) for 3 hr before separating through ultracentrifugation on a 5–40% glycerol gradient. (B) As in (A), reconstituted complex was incubated on ice with equimolar concentration of sNASP (competition assay) for 3 hr before ultracentrifugation. H3 elutes with Imp5, whereas sNASP elutes in its separate fraction. (C) As in (B), but adding Ran in equimolar concentrations to sNASP and Imp-H3. Ran in its purified state is unable to bind to Imp5 and compete with H3. (D) As in (B), but adding RanGTP in equimolar concentrations to sNASP and Imp5-H3 complex. In this instance, RanGTP associates with Imp5 and displaces H3, which co-elutes with its chaperone sNASP. Asterisks indicate an Imp5 degradation product.