Figure 2. Screening of DNA-damaging agents to extract the drugs activating the STING pathway in KL cells.
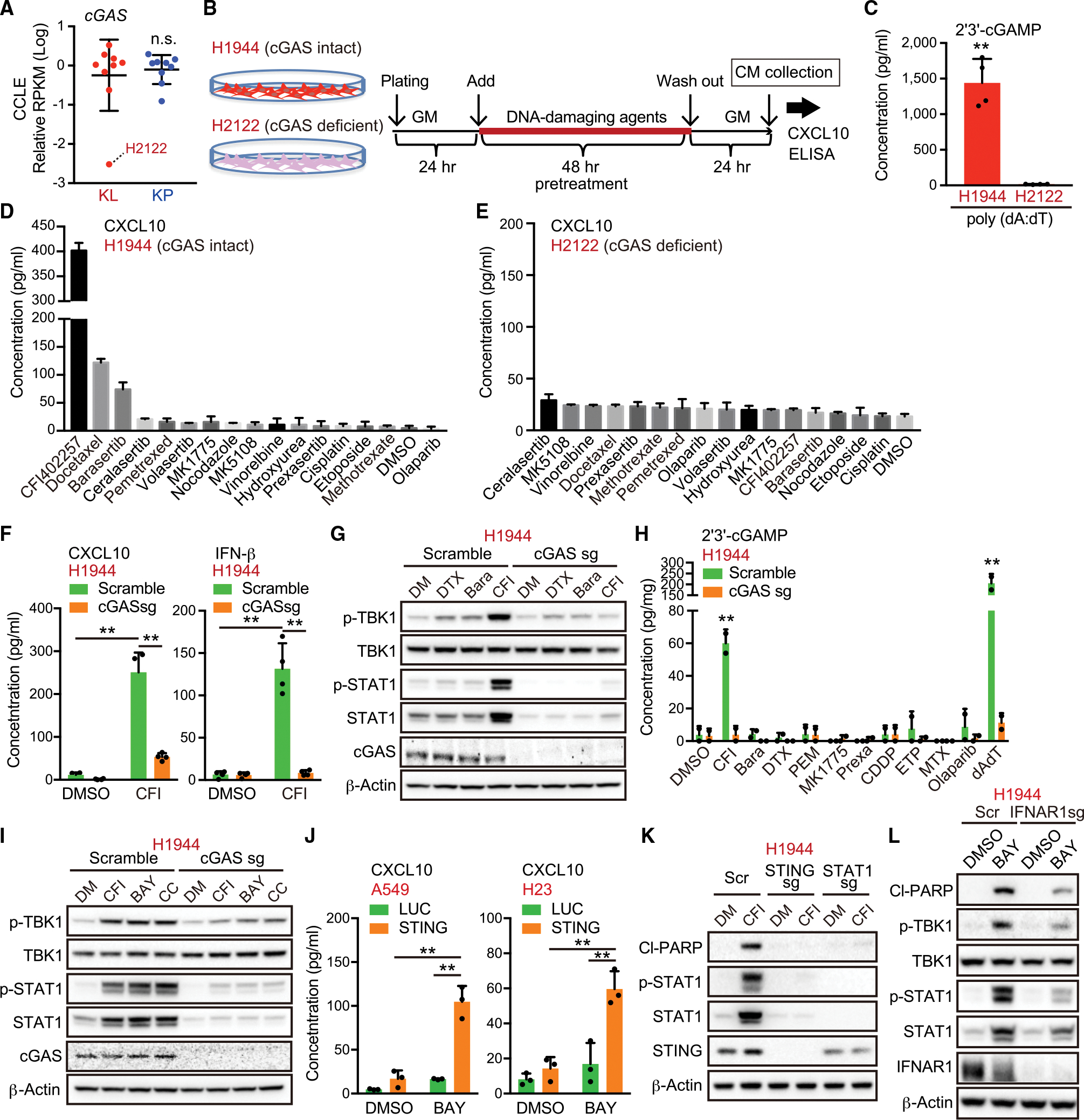
(A) Relative RPKM values of cGAS in KL and KP cells from CCLE.
(B) Schedule of drug treatment for the screening. GM, growth medium; CM, conditioned medium.
(C) Intracellular 2′3′-cGAMP levels in H2122 or H1944 cells treated with 0.5 μg/mL poly (dA:dT) (n = 4).
(D and E) Enzyme-linked immunosorbent assay (ELISA) of human CXCL10 in CM derived from H1944 (D) or H2122 (E) cells treated with the indicated DNA-damaging agents in accordance with the schedule for the screening (n = 2).
(F) ELISA of human CXCL10 or IFN-β levels in CM derived from H1944 cells transduced with the indicated vectors, treated with 200 nM CFI-402257 (n = 4).
(G and H) Immunoblot (IB) of the indicated proteins (G), or intracellular 2′3′-cGAMP levels (H), in H1944 cells transduced with the indicated vectors, and treated with the indicated DNA-damaging agents (n = 2). Bara, barasertib; CDDP, cisplatin; CFI, CFI-402257; DTX, docetaxel; ETP, etoposide Pr exa, prexasertib; MTX, methotrexate; PEM, pemetrexed.
(I, K, and L) IB of the indicated proteins in H1944 cells transduced with the indicated vectors, and treated with 200 nM CFI-402257, 100 nM BAY-1217389, or 250 nM CC-671.
(J) ELISA of human CXCL10 levels in CM derived from H1944 cells transduced with the indicated vectors, and treated with 100 nM BAY-1217389 (n = 3).
All quantitative data are represented as mean ± standard deviation; p values were calculated by unpaired two-tailed Student t test (A, C, and H), or two-way analysis of variance followed by Sidak’s post hoc test (F, J), **p < 0.01. See also Figure S2 and Table S1.
