Figure 6. Sequential combination therapy with MPS1 and DNMT inhibitor enhances intra-tumoral T cell infiltration in syngeneic murine KL model.
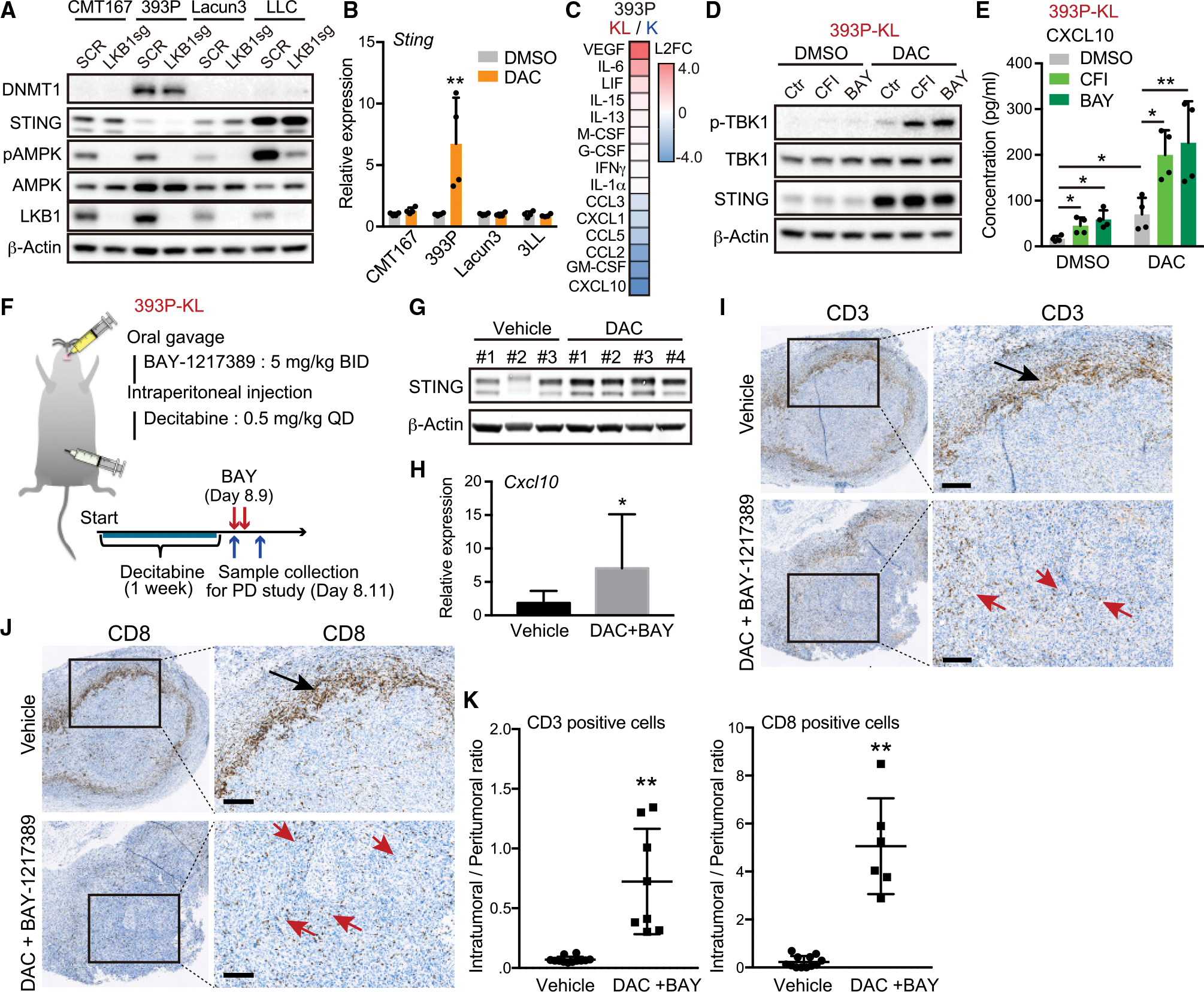
(A) Immunoblot (IB) of the indicated proteins in murine lung cancer cells transduced with the indicated vectors.
(B) Quantitative RT-PCR of Sting in murine lung cancer cells treated with 100 nM DAC for 5 days (n = 4).
(C) Heatmap of cytokine profiles in CM derived from 393P-K or 393P-KL cells. Scores = log2 fold change (393P-KL/393P-K). Cytokines indicating log2 fold change (L2FC) > 0.2 or L2FC < −0.2 are shown in the heatmap.
(D and E) IB of the indicated proteins (D), or Enzyme-linked immunosorbent assay (ELISA) of mouse CXCL10 levels in CM derived from 393P-KL cells (E) treated with the indicated drugs (100 nM DAC, and/or 200 nM CFI or 100 nM BAY) in accordance with pretreatment schedule (n = 4).
(F) Schematic of pharmacodynamics study with MPS1 and DNMT inhibitor in syngeneic murine KL model.
(G and H) IB of the indicated proteins (G), or quantitative RT-PCR of Cxcl10 (H) in tumor tissues derived from mice treated with the indicated drugs (each group, n = 4).
(I–K) Representative CD3 (I) or CD8 (J) IHC images and quantitative analysis (K) from 393P-KL tumors treated with vehicle or combination of DAC and BAY-1217389. Arrows highlight peri-tumoral localization (black) and intra-tumoral localization (red) of CD3+ or CD8+ T cells. QuPath (see STAR Methods) was used to quantify CD3+ or CD8+ T cell infiltration (n = 6). Scale bar, 200 μM.
All quantitative data are represented as mean ± standard deviation; p values were calculated by an unpaired two-tailed Student t test (B, H, and K), or two-way analysis of variance followed by Sidak’s post hoc test (E). *p < 0.05, **p < 0.01. See also Figure S6.
