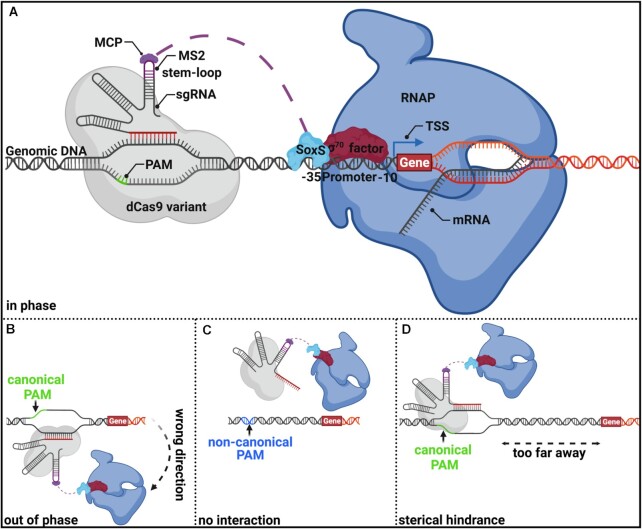
Figure 1.
General composition and limitation of a CRISPRa-construct consisting of a dead Cas9 protein variant (dCas9, dxCas9 (3.7), or dSpRY), with an MS2 coat protein (MCP) fused to transcriptional activator SoxS. (A) CRISPRa-construct is recruited to a particular upstream promoter region within the up-regulative window of approx. 70–90bp upstream of the TSS. This target site must be adjacent to a canonical PAM sequence (NGG) in the case of conventional Cas9 and allow proper spatial orientation of the transcriptional activator in phase with the target promoter. SoxS interacts with the conserved region 4 of the σ factor 70 family (in this example with σ70 in red) as well as with the α subunit of the core RNAP (blue) and build thereby the RNAP holoenzyme, which initiates transcription of the corresponding target promoter. (B) No transcriptional up-regulation will occur when targeting sites within the up-regulative window but with imperfect spatial orientation out of phase relative to the promoter. This is most likely attributed to the helical structure of the DNA, reoccurring every 10–11 bp. The graphic depicts the situation of a 5bp shift from the ideal target site. (C) Non-canonical PAM sites adjacent to the target site cannot be recognized by conventional Cas9; thus, no interaction of the CRISPRa construct and the target site will occur. (D) Targeting sites outside the up-regulative window sterically hinder the interaction of the recruited RNAP holoenzyme with the promoter.