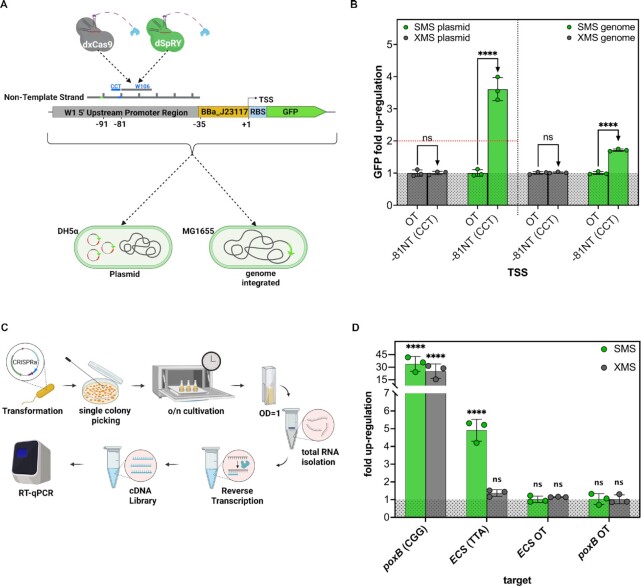
Figure 3.
SMS is functional at non-canonical PAMs during transgene and endogenous gene up-regulation. (A) To verify transgene up-regulation with SMS at non-canonical PAMs, we used E. coli MG1655 with genome-integrated GFP which is under the control of promoter BBa_J23117. For the up-regulation of transgenic GFP, we used XMS and SMS and targeted position -81 bp which is adjacent to non-canonical CCT. For comparison, we also targeted the original GFP plasmid on the same position in E. coli-DH5α. (B) SMS mediated up-regulation on plasmid (3.61-fold) and genome-integrated GFP (1.71-fold) resulted in considerable higher GFP levels compared to its off-target controls (OT), while XMS does not. All measurements were performed in n= 3 biological replicates (C) Afterwards, we performed RT-qPCR to verify endogenous up-regulation propensities with XMS and SMS at non-canonical PAMs. We chose endogenous ECS as a model protein and targeted it at position -80NT which is adjacent to a TTA PAM. For this purpose, we transformed E. coli-DH5α with CRISPRa plasmids and inoculated them into LB-medium with appropriate antibiotics, followed by incubation overnight. On the next day, we diluted the suspension 1:100 and let them grow until an OD of 1. Total RNA was then isolated and reverse transcribed followed by RT-qPCR measurement. (D) RT-qPCR studies at position -80 NT revealed statistically significant up-regulation of ECS with SMS (4.91-fold) and no up-regulation with XMS (1.37-fold). The positive control poxB could be up-regulated by both, SMS and XMS, which verifies their general functionality. All RT-qPCR datapoints were performed in n= 3 biological replicates, measured in technical replicates according to MIQE guidelines. Two-way ANOVA has been performed for plate-reader and one-way ANOVA for RT-qPCR experiments, confirming the statistical significance up-regulation of SMS. Results of all statistical analyses are provided in the Supplementary File Statistics. ****P< 0.0001. OT = off-target control