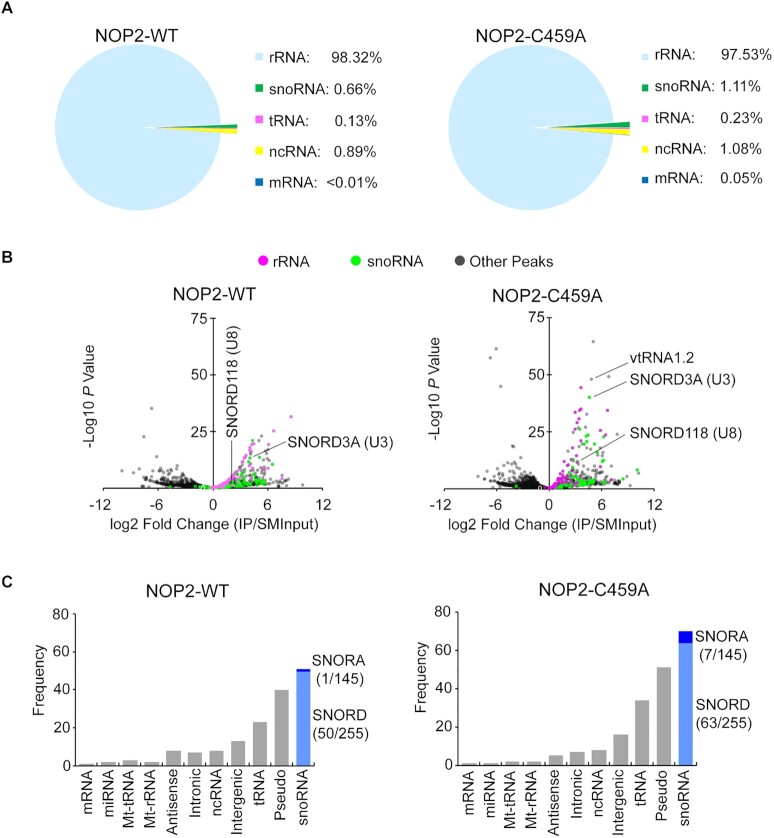
Figure 2.
Human NOP2/NSUN1 binds to rRNA and snoRNAs. (A–C) HEK293T cells expressing FLAG-tagged NOP2/NSUN1 WT (non-covalently bound to substrate) or C459A mutant (covalently bound to substrate) were subject to CLIP-sequencing analysis (two biological replicates for each sample). CLIP-sequencing reads were aligned to human genome (hg38) and repetitive elements families. CLIP peaks were called by Peakachu. (A) Percentage of miCLIP mapped peak regions in each indicated RNA category with cut-offs of adjusted significance P values < 0.05 and fold change over size-matched input (SMInput) > = 2. (B) Volcano plots of all peaks detected by Peakachu. Each dot represents a peak either enriched in IP (log2 fold change > 0) or SMInput samples (log2 fold change < 0). (C) Number of CLIP peaks with cut-offs of adjusted significant P values < 0.05 and fold change over SMInput ≥2 in each indicated RNA category. C/D box and H/ACA box snoRNA are shown in light and dark blue, respectively. Number of snoRNAs found in CLIP peaks / total number of snoRNAs in the indicated subfamily are shown in parentheses.