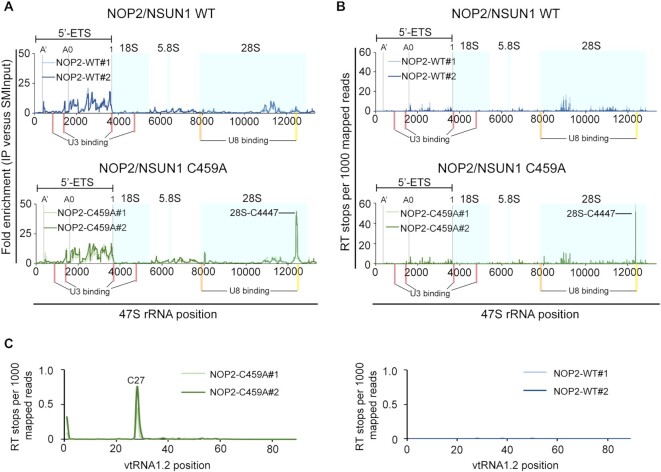
Figure 3.
Human NOP2/NSUN1 binds to the rRNA 5′-ETS region and crosslinks on 28S rRNA at position C4447. (A, B) miCLIP-sequencing data were aligned to human 47S pre-rRNA (NR_046235) and the mapping information was retrieved using Samtools. The location of mature 18S, 5.8S and 28S rRNAs are marked with light blue shadow boxes. A’, A0 and 1 cleavage sites on 5′-ETS are marked with gray lines. Red bars indicate the location of U3 snoRNA binding sites (101). The orange bar indicates the location of U8 snoRNA binding site based on sequence homology and accessibility measurements (103). The yellow bar indicates the location of the U8 snoRNA binding site found by RNA duplex mapping (106). (A) Plot of sequencing depth-normalized reads ratio of IP over size-matched input (SMInput) on the 47S rRNA. (B) Plot of reverse transcription (RT) stops on the 47S rRNA normalized by sequencing depth. RT stops were assigned at the start (+1) sites of the Read1 sequence from NOP2/NSUN1 WT and C459A IP samples. (C) Plot of reverse transcription (RT) stops on vtRNA1.2 normalized by sequencing depth. RT stops were assigned at the start (+1) sites of the Read1 sequence from NOP2/NSUN1 WT and C459A IP samples. (B, C) Plotted data is represented for each independent biological replicate for NOP2/NSUN1 WT (blue lines) and C459A mutant (green lines).