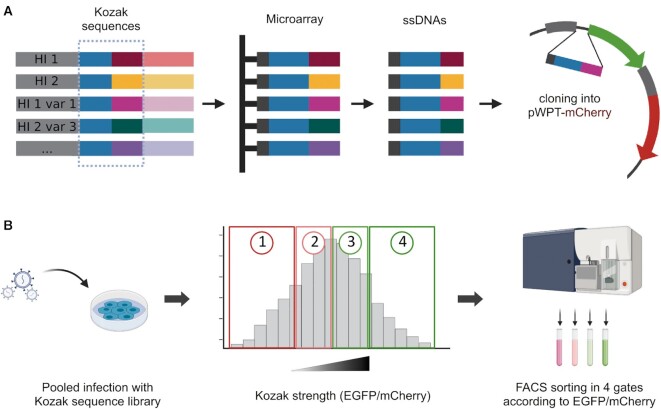
Figure 2.
Schematic workflow of library generation and selection screening for Kozak strength. (A) The Kozak variants were designed as oligonucleotides bearing the overhangs to be cloned in the destination vector. The oligos were synthesised on a custom microarray. The library was cloned in place of the EGFP Kozak sequence in a bicistronic reporter vector. (B) The Kozak sequence library was used to transduce HEK293T cells. Transduced cells were sorted according to their EGFP/mCherry ratio as a measure of Kozak strength. The four gates were drawn so that each gate contained 25% of the total population.