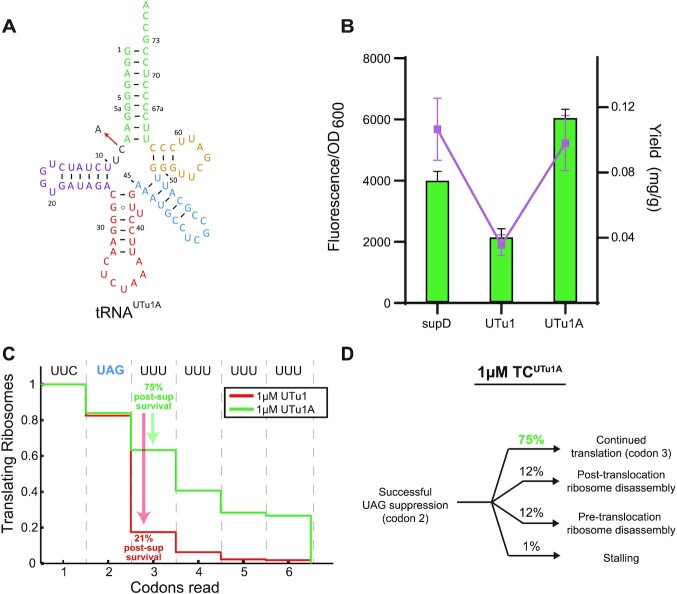
Figure 5.
Single nucleotide mutation enhances translation processivity. (A) Cloverleaf structure of tRNAUTu1A showing the single nucleotide C to A change from tRNAUTu1 by the red arrow. (B) sfGFP fluorescence readthrough assay (green bars) and GPx1 protein yields (purple dots) demonstrate the increased processivity of tRNAUTu1A compared to tRNAUTu1. Fluorescence reads are shown as an average of four biological replicates (± standard deviation) and protein yields are an average of two biological replicates (± standard deviation). (C) Codon survival curve plotting the fraction of translating ribosomes as a function of number of codons translated highlights the post-suppression survival (percent of codon 2 ribosomes that survive codon 3) advantage in the presence of 1 μM tRNAUTu1A (green, n = 300) over 1 μM tRNAUTu1 (red, n = 223). (D) Pathway map of ribosomes that successfully suppressed a UAG stop codon in the presence of 1 μM tRNAUTu1A.