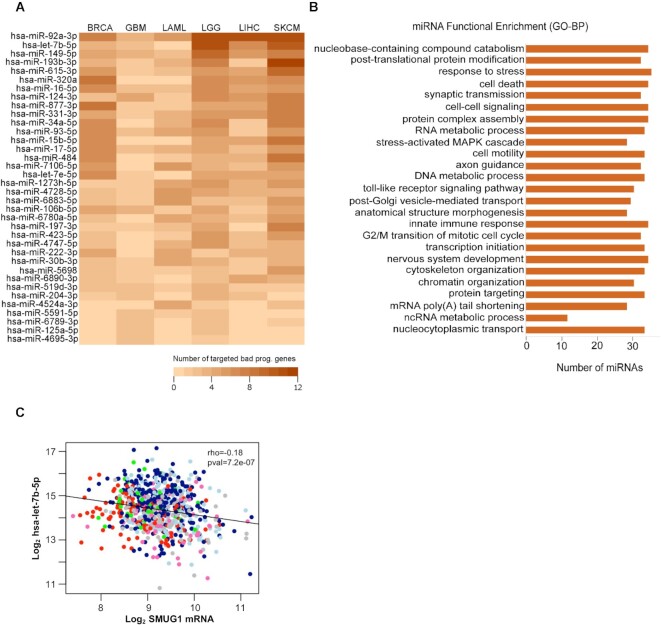
Figure 3.
Identification of upstream miRNA regulators of SMUG1 interacting proteins in TCGA datasets. (A) Heatmap of the miRNAs (n = 36) regulating the highest number of SMUG1 interacting proteins in BRCA, GBM, LAML, LGG, LIHC and SKCM datasets. Heatmap is ordered, from highest to lowest, according to the total number of proteins in each dataset which is regulated by corresponding miRNAs on y-axis. Color scale is showing the number of genes targeted by those miRNAs in each dataset, lightest as lowest and darkest as highest number of genes. (B) Functional annotation of top 10 miRNAs of all datasets (n = 36) presented in (A) based on Gene Ontology—Biological Process terms (P < 0.05). (C) Scatterplot showing SMUG1 mRNA expression (x-axis) versus hsa-let-7b-5p expression (y-axis) for breast tumor samples of the TCGA–BRCA cohort (n = 747). Tumors are color-coded according to the PAM50 molecular subtype; dark blue = luminal A (LumA; n = 267), light blue = luminal B (LumB; n = 49), red = basal-like (basal; n = 115), pink = Her2-enriched (HER2; n = 62), green = normal-like (n = 49), gray = subtype not available (n = 106). Spearman′s rho and the associated P-value are indicated.