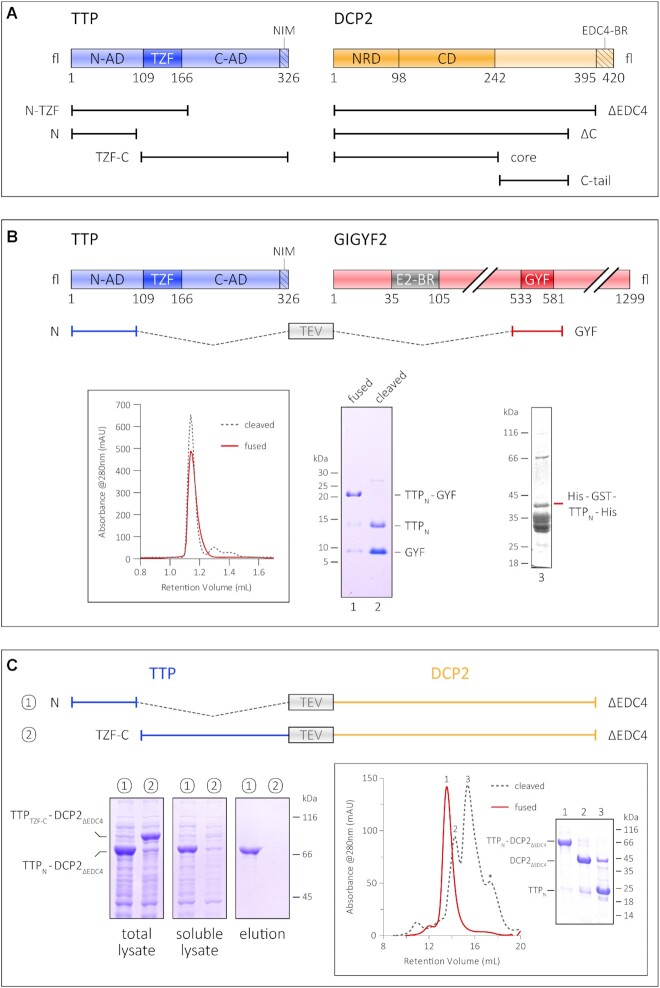
Figure 1.
Interaction of the TTP N-terminal activation domain with GIGYF2 and DCP2. (A) Schematic representation of the domain organization of TTP and DCP2 and the constructs used in this study. The central tandem zinc finger domain of TTP (dark blue) is flanked by the unstructured N- and C-terminal activation domains (N-AD and C-AD, respectively) on either side (light blue). The NOT1-interacting motif (NIM) at the very C-terminus of TTP is indicated by the hatched box. The structured core of DCP2 (dark orange) comprises its N-terminal regulatory domain (NRD) and its catalytic Nudix domain (CD), and is followed by a long, unstructured C-terminal region (light orange), the last 25 residues of which harbour a binding motif for EDC4 (EDC4-BR, indicated by the hatched box). (B) The GYF domain of GIGYF2 restores proteolytic stability of the TTP N-terminal activation domain (TTPN). A schematic representation of GIGYF2 indicates the position of the GYF domain (dark red) in the primary structure of the protein, with respect to other binding motifs, such as the eIF4E2-binding motif (in grey, denoted by E2-BR). The fusion polypeptide of TTPN and GYF, separated by a linker sequence bearing a TEV protease cleavage site, is shown. Analytical SEC and the corresponding SDS–PAGE analysis of peak fractions depict the stability of TTPN in complex with GYF. The retention volume of the single polypeptide (fused) is identical to that of non-covalently linked complex (cleaved, referring to cleavage by TEV protease), indicating formation of a stable complex. TTPN in complex with GYF is stable even upon TEV cleavage of the fusion polypeptide. In contrast, fusion of a non-interacting protein such as GST does not rescue the proteolytic instability of TTPN. (C) Schematic representation of the TTP–DCP2 fusion polypeptides, designed similarly to the TTPN–GYF fusion described above, and comparison of the stability of TTPN–DCP2ΔEDC4 with that of TTPTZF–C–DCP2ΔEDC4 by Ni2+-affinity pulldowns. Both fusion proteins are well expressed, but only TTPN–DCP2ΔEDC4 can be isolated by Ni2+-affinity purification, indicating that fusion of DCP2 rescues the behaviour of TTPN but not that of TTPTZF-C. The left, middle and right panels indicate total lysate, soluble lysate and elution, respectively. Total lysate and soluble lysate refer to samples collected before and after centrifugation of the cell suspension after sonication. Inset: analytical SEC and the corresponding SDS–PAGE analysis of peak fractions of the intact TTPN–DCP2ΔEDC4 fusion and of the protein cleaved with TEV protease. Cleavage of the fusion yields proteolytically stable TTPN, although it is not stably associated with DCP2ΔEDC4 in SEC.