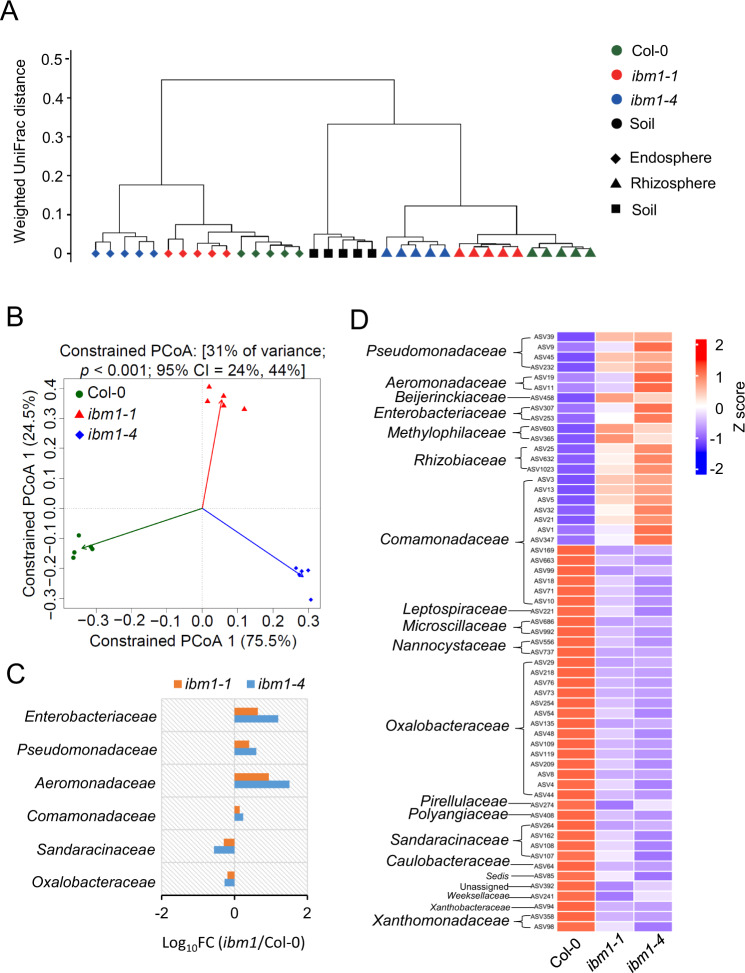
Fig. 1. IBM1 dysfunction reshapes Arabidopsis root microbiome.
A The hierarchical clustering of the weighted UniFrac distances between samples highlights the importance of compartment and genotype to microbiome. B The PCoA analysis of all ASVs separates the ibm1 mutants from the wild-type plants (Col-0) within the endosphere compartment. C The ibm1 mutations resulted in altered (FDR < 0.05 for both the ibm1-1 vs Col-0 and the ibm1-4 vs Col-0) RA in 6 bacteria families within the endosphere microbiome. The Log10 fold changes of mutant vs wild type are shown as horizontal bars. D The ibm1 mutations resulted in altered (p < 0.05) RA in 59 endosphere ASVs, with 38 (64%) becoming less enriched in the mutants compared to the wild type. The notes on the left of the heatmap show the taxonomy of each ASV at the family level.