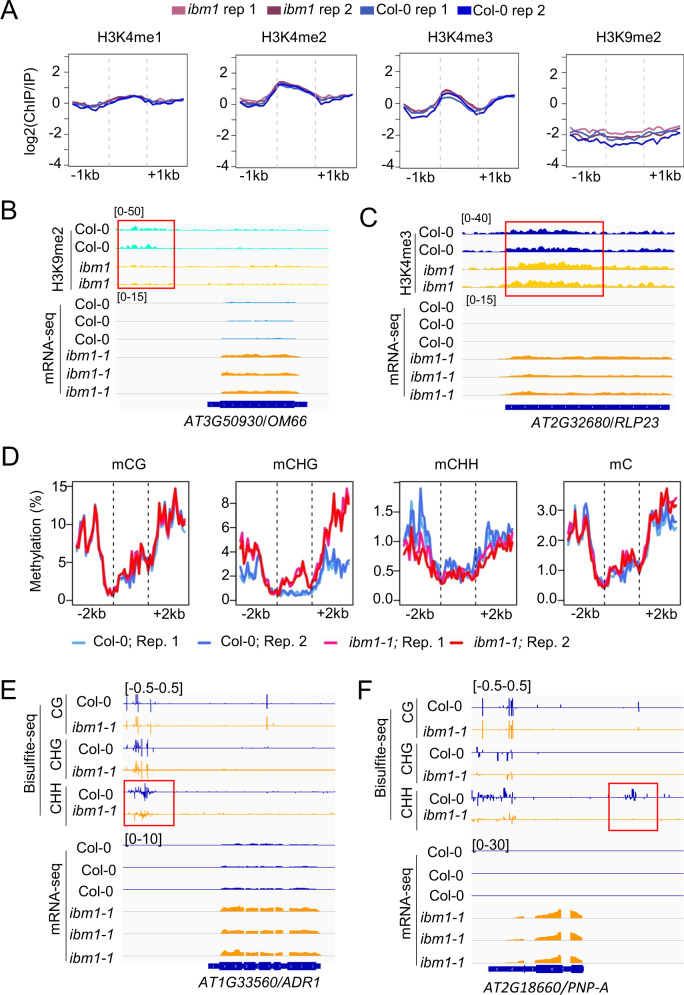
Fig. 3. Epigenome analyses identified ibm1 mutation-induced changes permissive for transcription of important defense genes.
A The overall patterns of H3K4me1, H3K4me2, H3K4me3, and H3K9me2 levels at the 178 defense DEGs and the vicinity regions (up- and downstream of 1 kb). The original ChIP-seq data were downloaded from DDBJ (DRA005154) as generated previously (22). B IBM1 dysfunction decreases H3K9me2 level at the gene promoter of OM66 and increases the mRNA level of OM66. Snapshots from ChIP-seq and RNA-seq are shown. The red box indicates the region with altered H3K9me2 levels. C IBM1 dysfunction increases H3K4me3 level at the gene body region of RLP23 and increases the mRNA level of RLP23. D The overall patterns of DNA methylation levels at the 178 defense DEGs and the vicinity regions (up- and downstream of 2 kb). E IBM1 dysfunction decreases DNA methylation level at the promoter region of ADR1 and increases the mRNA level of ADR1. Snapshots from whole-genome bisulfite sequencing and RNA-seq are shown. The red box indicates the gene promoter region with altered CHH methylation levels. F IBM1 dysfunction decreases DNA methylation level at the promoter region of PNP-A and increases the mRNA level of PNP-A.