Figure 7.
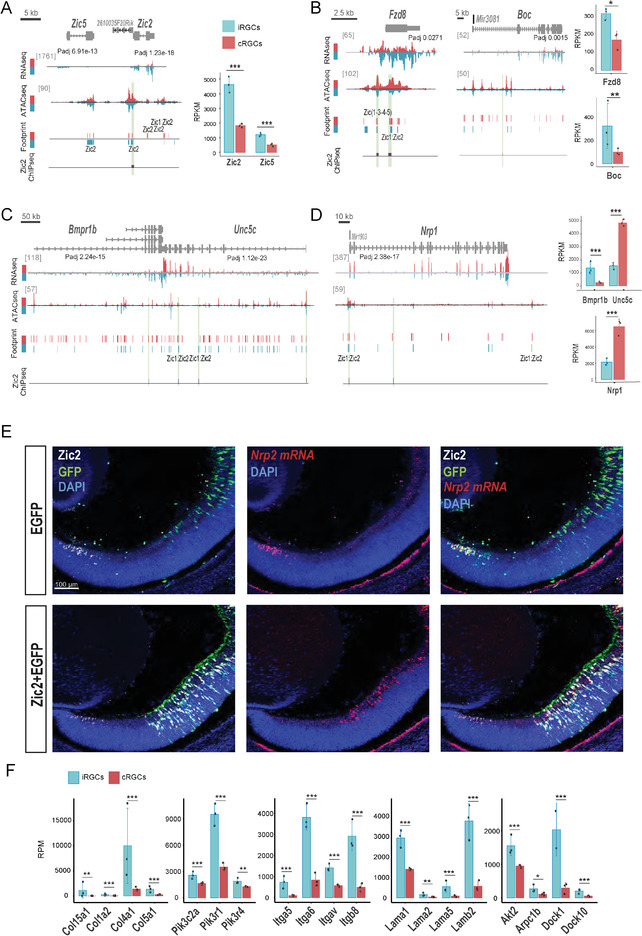
Known and novel differentially expressed genes (DEGs) in ipsilateral retinal ganglion cell (iRGC) or contralateral retinal ganglion cells (cRGCs). A) Genomic snapshots of RNA‐seq, ATAC‐seq, digital footprints, and Zic2 ChIP‐seq in iRGC and cRGCs at the Zic2/Zic5 locus. Zic2 transcripts are highly expressed in iRGCs. The Zic2/Zic5 locus contains several potential Zic1:Zic2 footprints. Actual binding of Zic2 to the site located in the TSS of Zic2 was confirmed by ChIP‐Seq (green band). Graph at the right represents transcripts expression for Zic2 and Zic5 (in reads per million, RPM) obtained from iRGC or cRGCs RNA‐seq experiments. B–D) Genomic snapshots of RNA‐seq, ATAC‐seq, digital footprints in cRGC (red)/iRGCs (blue), and Zic2 ChIP‐seq in retina/spinal cord at loci that encode for receptors differentially expressed in iRGCs and cRGCs. Most of these loci contain potential Zic1:Zic2 footprints and some of them were confirmed by ChIP‐seq (green band). E) Coronal retinal sections from E16.5 embryos electroporated at E13.5 with plasmids encoding for EGFP‐control plasmids (upper panels) or Zic2‐ plus EGFP‐encoding plasmids (bottom panels). Immunostaining for Zic2 combined with in situ hybridization for Nrp2 mRNA demonstrate that Nrp2 is highly and specifically expressed in endogenous Zic2 positive cells in the peripheral retina but not in targeted cells expressing EGFP. However, when Zic2 is ectopically expressed, Nrp2 mRNA levels are highly increased. Scale bar: 100 µm. F) Graphs represent transcripts expression (in RPM) obtained from iRGC or cRGCs RNA‐seq experiments corresponding to genes encoding for different integrins (‐t test, ***Padj < 0.001).
