Figure 2.
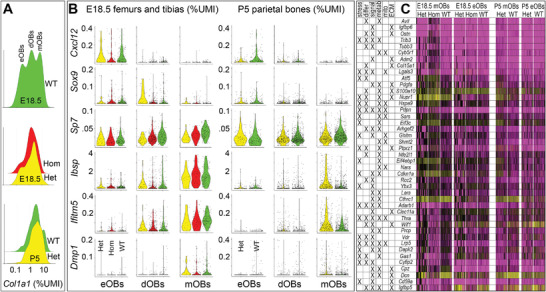
Transcriptomic signatures of osteoblasts (OB) differentiation and function. A) Histograms of Col1a1 expression in E18.5 long bones (top 2 panels) and P5 parietal bones (bottom panel). Hereafter mRNA expression is reported as percent of the total transcripts in the cell (%UMI). Based on distinct peaks in the Col1a1 histogram fromWT E18.5 bones, we separated three subpopulations of OBs with Col1a1 < 0.75%UMI, 0.75 ≤ Col1a1≤3.5%UMI, and Col1a1 > 3.5%UMI in all genotypes and tissues. B) Expression of osteoblast differentiation markers. Based on relative expression of Cxcl12, Sox9, Sp7, Ibsp, Ifitm5, and Dmp1, we classified the first subpopulation as early (eOBs), the second one as differentiating (dOBs), and the third one as mature (mOBs) osteoblasts. C) Heat maps of genes significantly affected by the G610C mutation in mOBs from both E18.5 long bones (Figure 1) and P5 parietal bones (4 Het, 954 OBs; 4 WT, 1521 OBs). The color change from magenta to black to yellow corresponds to increasing gene expression. All genes satisfying the following criteria are shown: i) statistically significant effect of the mutation both in Hom E18.5 mOBs and Het P5 mOBs (p < 0.05, Wilcoxon test with Bonferroni correction); ii) at least twofold change in Hom versus WT E18.5 mOBs; iii) reproducible effect of the mutation in two independent experiments with P5 OBs; and iv) involvement of the gene in at least two processes essential for OB function (marked by X). The following essential processes annotated in the Gene Ontology (GO) database were chosen for the analysis: cell death, stress, transcription, translation (stress); cell differentiation (differ); cell signaling (signal); metabolism (metab); mitochondrial function (mito); and extracellular matrix (ECM) synthesis/function (ECM). Cautionary notes: a) The Bonferroni correction for multiple comparisons may prevent detection of some genes affected by the mutation (particularly when low expression limits accurate mRNA quantification by scRNASeq). b) While useful for initial bioinformatic analysis, GO annotations may be misleading since they are incomplete and do not require validating experiments.
