Figure 6.
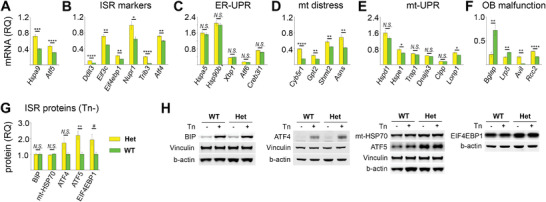
Relative quantity (RQ) of key pathway/function markers in cultured primary OBs. A‐F) RNASeq quantification of mRNA relative to geometric mean of housekeeping genes (Actg1, Actb, Mrfap1, and Sdha). G) Cell lysate protein quantification in Het relative to average WT values by Western blotting. H) Representative blots of the proteins and corresponding loading standards (each panel shows a separate blot). Short (3–5 h) treatment with 10 µg mL−1 tunicamycin (Tn) was used for validating effects of canonical, acute UPR on BIP and ATF4 (not expected to produce noticeable effects on mt‐HSP70, ATF5, and EIF4EBP1). A) Hspa9 and Atf5. B) ISR genes. C) Endoplasmic reticulum (ER) UPR genes (Hspa5, Hsp90b1, Xbp1, Atf6) and Creb3l1. D) Markers of mitochondrial distress. E) Markers of mt‐UPR. F) Markers of cellular malfunction: Bglap – OB differentiation, Lrp5 – Wnt signaling, Avil – cytoskeleton and cell motility, Rcc2 – cell cycle. Error bars show standard error of the mean in six replicates (mRNA) and 6–12 replicates (protein). N.S. = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. More than 20% (see Table S1, Supporting Information) changes in relative expression with p < 0.05 were considered statistically significant. Two‐tailed, heteroscedastic t‐test and Mann–Whitney U‐test produced consistent significance estimates for most markers, with the only exception of p = 0.03 in t‐test and p = 0.1 in U‐test indicated by # (EIF4EBP1 in G). For mRNA analysis, six wells per genotype were examined (3 at 14 and 3 at 21 days after seeding) from two separate cell preparations (two wells per genotype in the first and four wells per genotype in the second experiment). Similarly, six wells per genotype were examined by Western blotting in two separate cell preparations of three wells per genotype. The variation between the experiments was the same as between different wells within the same experiment. Technical variation between different Western blot lanes was comparable to or larger than the variation between different biological samples.
