Figure 1 – Cytokines and cytokine signaling altered in CD11chi lupus prone NZM2410 mice expressing different forms of ERα.
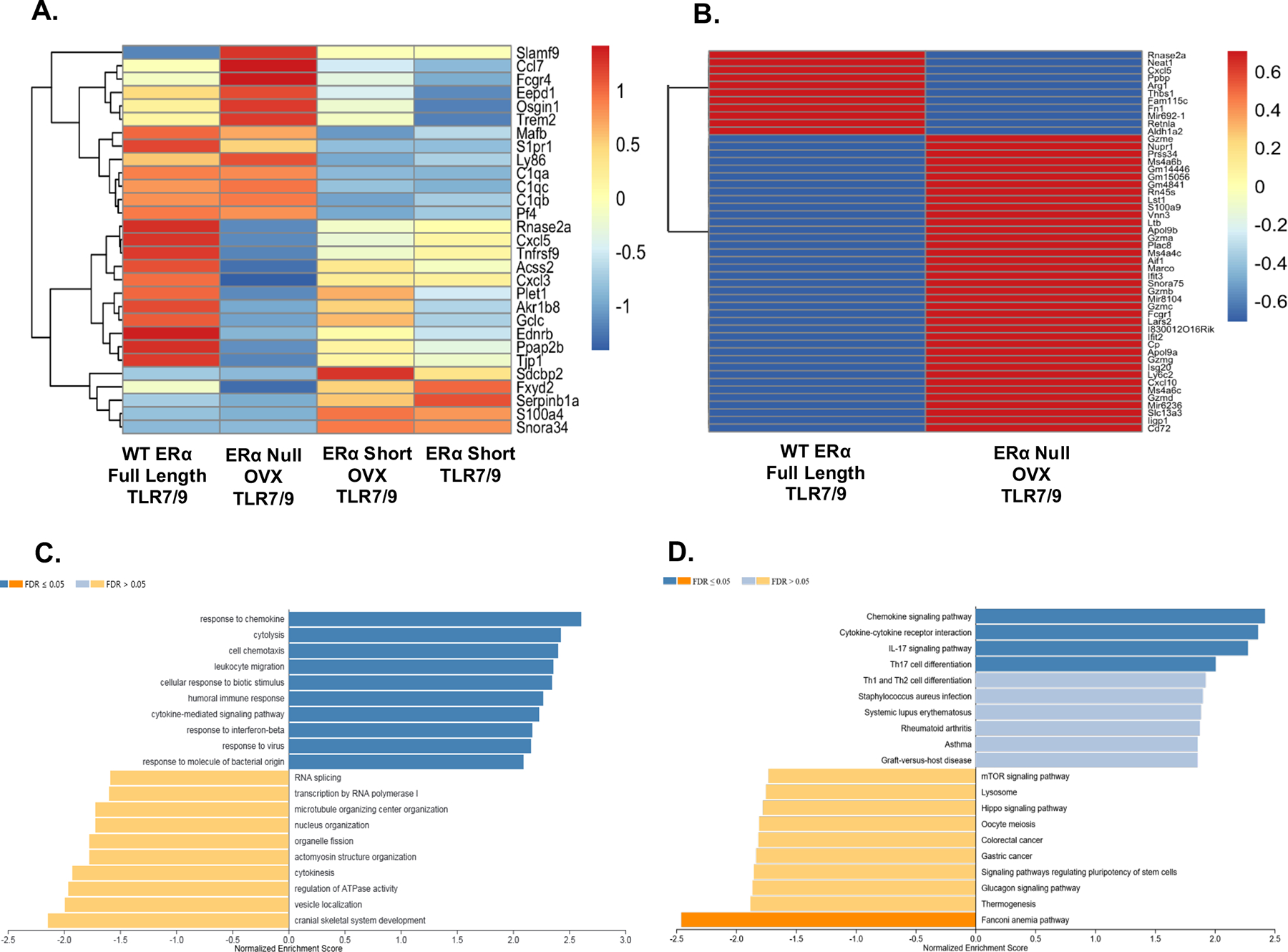
Heatmaps revealing differentially expressed genes (DEGs) in CD11c hi cells isolated from lupus mouse strains with different forms of ERα. CD11chi cells were derived from murine bone marrow cultures with GM-CSF and IL-4. Panel A depicts DEGs across all four strains of mice (with and without ovariectomy). Values are in log10-based FPKM. All cells were stimulated with TLR7/9 and hormonal differences were accounted for in the analysis. Panel B shows the DEGs across WT and ERα null only. The same parameters used in Panel A apply to Panel B. The bar plot in Panel C depicts the GO analysis results for genes between WT and ERα null that have an absolute fold change over 1 and included a total of 1912 genes. Panel D represents the results of the KEGG pathway analysis using the same set of genes as in Panel C. For Panels C and D the WT group was used as the reference gene set.
