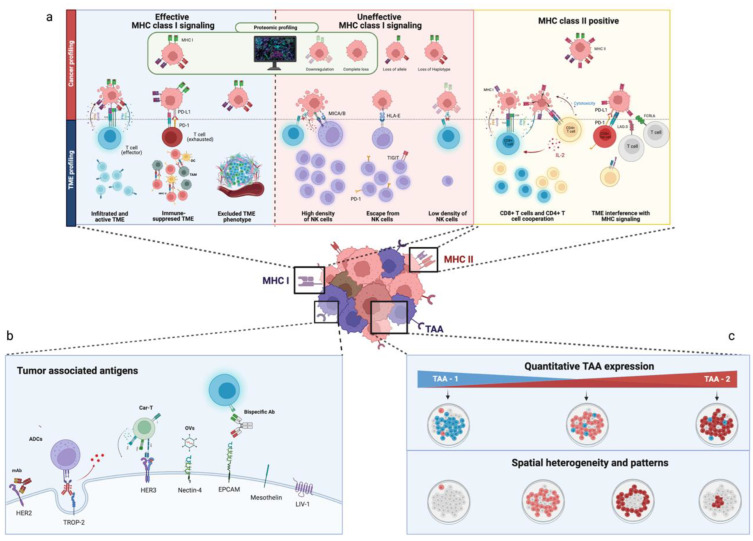
Figure 1.
Multiplexed spatial protein profiling of breast cancer antigenicity. (a) Major histocompatibility complex (MHC) class I and II cancer (upper quadrants) and TME profiling (lower quadrant). MHC class I alterations that can be accurately characterized by proteomic profiling are underlined in green. In the blue box: Cancer cells with effective MHC-I signaling can be killed by effector cells or survive despite MHC-I expression if, for example, T cells are present but exhausted, or the TME lacks T cell infiltration, as is the case in an immune-excluded TME. In the green box: Cancer cells lacking MHC class I can be killed by NK cells, but can escape immune-killing in a TME that lacks NK cell infiltration or employs countermeasures that limit NK cell-mediated killing, such as in HLA-E expression. In the yellow box: MHC class II cancer cells can be killed by the coordinated effort of both CD8+ T cells and CD4+ T cells. Nonetheless, an infiltration of LAG3+ and FCRL6+ TILs can interfere with MHC class II signaling and induce cancer cell survival. (b,c) Cancer profiling of tumor-associated antigens’ (TAAs) quantitative expression and spatial distribution. Figure 1b shows relevant TAAs that can be characterized through multiplexed in situ protein profiling and some therapeutics that could benefit from TAA-profiling; Figure 1c underlines the ability of multiplexed spatial protein profiling in evaluating the combined quantitative expression of different TAAs (e.g., TAA-1 and TAA-2) in the same sample and their spatial heterogeneity. Created with BioRender.com.