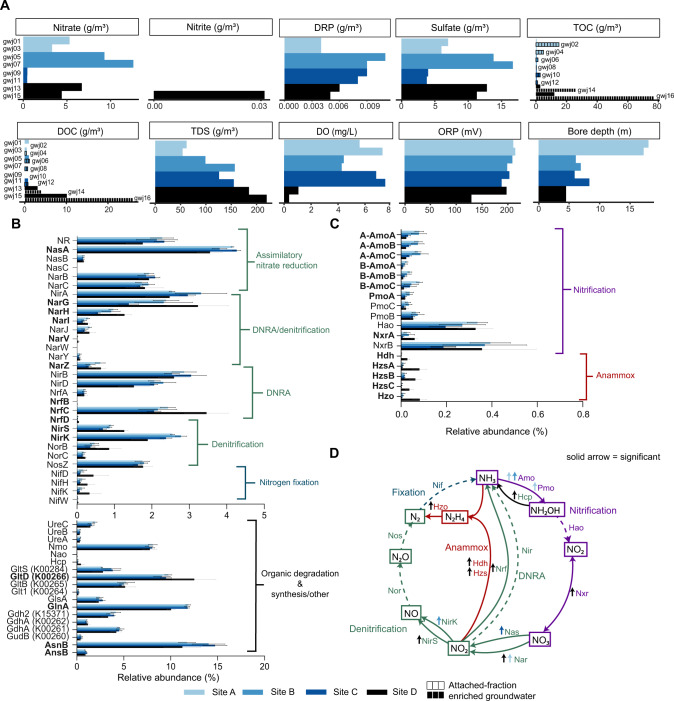
Fig. 1. Geochemistry and protein-coding sequences (based on reads) involved in nitrogen cycling that are significantly different among sites used for metagenomics.
A Bar plots showing geochemical data from groundwater samples, coloured according to site. Solid bar colour = groundwater samples. Grid lines = attached-fraction enriched groundwater. All samples from site D were characterized as dysoxic, although gwj15-16 contained 0.37 mg/L DO, which are near suboxic levels (i.e. <0.3 mg/L). For all samples shown, ammoniacal-N values were below detection. B, C Bar plots showing the abundance (average of four wells per site and standard deviation) of sequence reads encoding dissimilatory and assimilatory nitrogen-cycling proteins relative to all nitrogen-cycling processes. Predicted proteins that were statistically different between sites are bolded (y-axis). D Schematic of the nitrogen cycle displaying statistically significant differences between sites (LEfSe, Kruskal–Wallis test, p < 0.05). Solid lines depict pathways that were significantly more abundant across the sites, whereas dashed lines indicate no significant difference. Arrows indicate the site with significantly more genes. Abbreviations: A-Amo Archaeal Amo, B-Amo Bacterial Amo, Amo ammonia monooxygenase, Pmo Particulate methane monooxygenase, Hao hydroxylamine oxidoreductase, Nxr nitrite oxidoreductase, Nar nitrate reductase (dissimilatory), Nas nitrate reductase (assimilatory), NirK copper-containing nitrite reductase, NirS cytochrome cd1-containing nitrite reductase, Nor nitric oxide reductase, Nos nitrous oxide reductase, Nif nitrogenase (various), Hcp hydroxylamine reductase, Nir NADPH-nitrite reductase, Nrf nitrate reductase (associated with Nap), Hdh hydrazine hydrogenase, Hzs hydrazine synthase, Hzo hydrazine oxidoreductase.