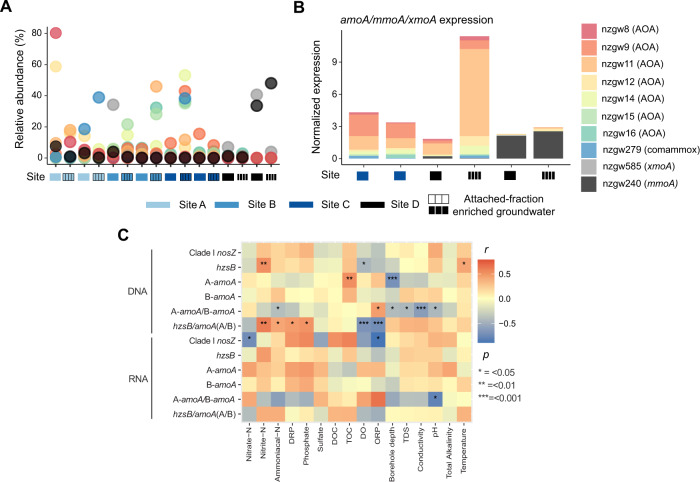
Fig. 6. Composition and transcriptional activity of ammonia-oxidizers, and relationship between ammonia-oxidizers, denitrifiers and anammox bacteria to geochemical and physical parameters.
A Relative abundance of each MAG capable of aerobic ammonia oxidation across the sites. B Stacked barplot showing modified transcripts per million, normalized to genome coverage, of amoA, pmoA and xmoA across sites (wells SR1, SR2, E1, N3). The 7 AOA are associated with 5 genera: Nitrosotenuis (nzgw11), UBA8516 (nzgw12–14), Nitrososphaera (nzgw16), Nitrosoarchaeum (nzgw8-9) and an unclassified genus in Nitrososphaeraceae (nzgw15). The xmoA is part of a gene cluster in MAG nzgw585, recovered from the dysoxic site and classified as Gammaproteobacteria genus Nevskia, which encodes a copper-containing membrane monooxygenase (CuMMO/xmoCAB). The alpha subunit had best hits to Polycyclovorans sp. SAT60 and gammaproteobacterial isolate MMS_B.mb.28 (85.71% amino acid identity, NCBI NR database). CuMMO catalyzes the oxidation of short-chain alkanes, ammonia or methane [103]. C Spearman’s rank correlations between the abundance (copies/L) of nitrogen-cycling genes (DNA, samples = 64) and transcripts (RNA, samples = 26 above detection limit) determined via ddPCR and geochemical parameters (Table S11). Significant correlations are indicated by * are based on Bonferroni adjusted p values (p).