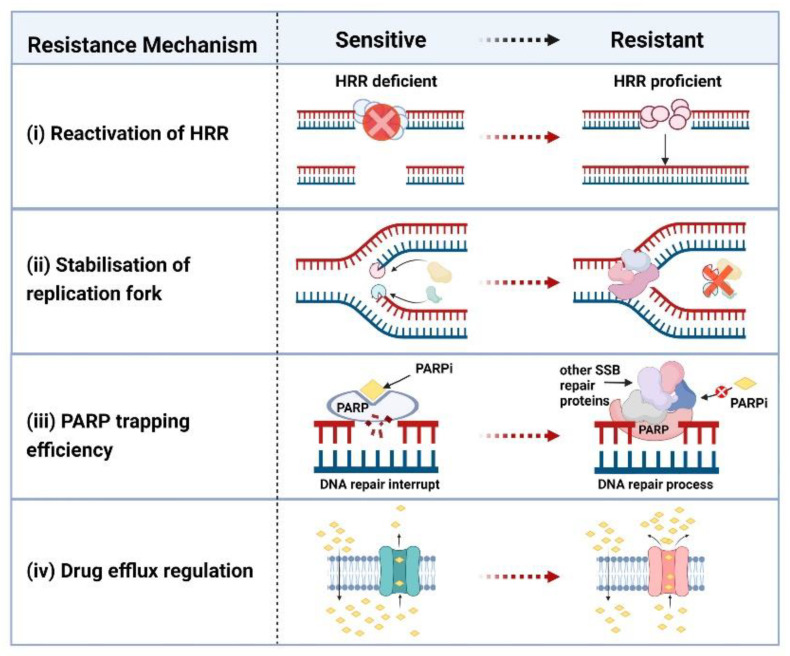
Figure 4.
Mechanisms of PARP inhibitor resistance. There are four generally accepted mechanisms of PARPi resistance. (i) Homologous Recombination (HRR) restoration. The synthetic lethality is based on HRR deficiency and PARP inhibition. When HR reactivates in cancer cells for reasons including the occurrence of a reversion mutation, DNA damage can be repaired by the HRR pathway, cancer cells survive and become resistant to PARP inhibitors (PARPis). (ii) Stabilisation of the replication fork. Some HRR-associated proteins also function to stabilise stalled DNA replication forks. In HRR deficient cells (sensitive to PARPi), stalled replication forks will be degraded by DNA nucleases (e.g., MRE11, MUS81). PARPi resistant cells protect the replication fork by inhibiting the recruitment of DNA nucleases, therefore maintaining genomic stability. (iii) A PARP mutation can affect the ability of PARPi to bind PARP, leading to a decrease in PARP trapping. Mutated PARP can recruit other DNA repair proteins to correct single-strand breaks, leading to PARPi resistance and cell survival. (iv) Increase in drug efflux. Some PARPis (e.g., olaparib) are substrates of the transmembrane glycoprotein MDR1 (multi-drug resistance protein 1), also known as p-glycoprotein 1. By upregulating the activities of MDR1, PARPi is transported out of the cancer cells. Created with Biorender.com.