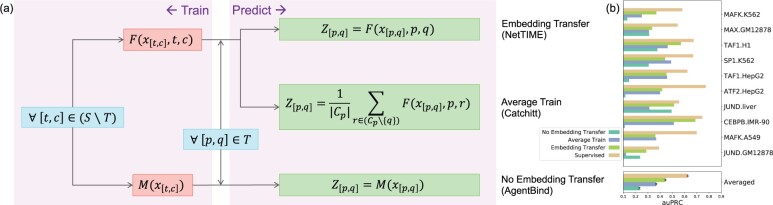
Fig. 5.
Comparison of different transfer learning strategies. (a) Detailed overview of the training scheme and prediction generation procedure using three transfer learning strategies implemented by NetTIME, Catchitt and AgentBind. and denote the particular TF (t or p) and cell type (c or q) combinations. S refers to the set of all conditions in our training dataset. T is the set of 10 leave-out conditions. Cp is the set of cell types that satisfy . is the input data from . is the per-base-pair binding probability predictions for . F and M can be any machine learning models. To avoid performance differences introduced by the model architecture, we use NetTIME model with TF and cell-type embeddings (F) and with no embedding (M). (b) Transfer predictions using different transfer learning strategies are generated for 10 leave-out TF and cell-type combinations within the training panels of TFs and cell types