Fig. 6.
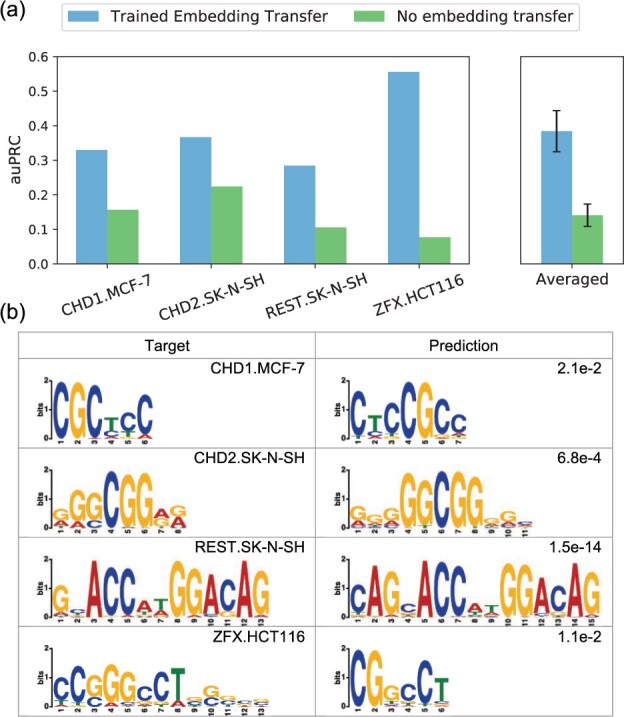
Transfer predictions to new TF and cell-type combinations beyond the training panels of TFs and cell types. (a) Transfer predictions using models trained with either TF and cell-type embedding vectors (Trained Embedding Transfer) or no trained embeddings (No embedding transfer). (b) Comparison of predicted TF binding motifs to those derived from target ChIP-seq conserved peaks. Predicted motifs are derived from Trained Embedding Transfer predictions. De novo motif discovery is conducted using STREME (Bailey, 2020) software. Motif similarity P-values shown in the top right corner of the Prediction column are derived by comparing predicted and target motifs using TOMTOM (Gupta et al., 2007)
