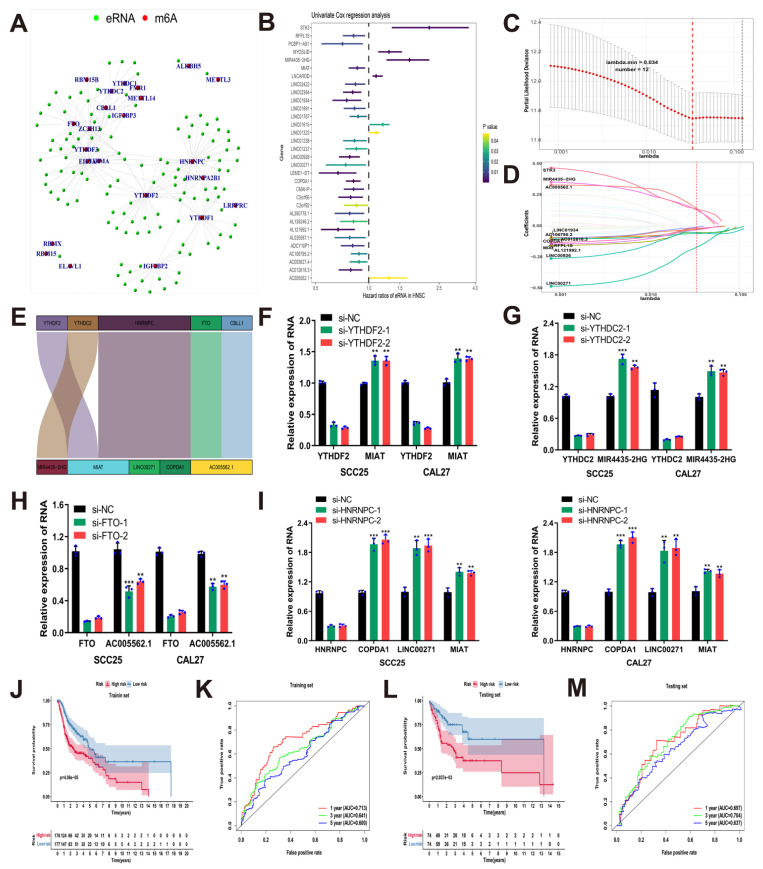
Figure 2.
Construction and validation of the risk model based on prognostic m6A-related eRNAs. (A) Network of m6A regulators as red nodes and 120 eRNAs as green nodes; (B) forest plot of univariate Cox regression analysis of 32 m6A-related eRNAs associated with OS; (C) selection of the optimal lambda in the LASSO model; (D) LASSO coefficient of the 12 m6A-related eRNAs; (E) Sankey diagram showing the relationship between m6A regulators and m6A-related eRNAs; (F) RT-qPCR were used to detect MIAT expression after transfection of siYTHDF2 in SCC25 and CAL27 cells; (G) RT-qPCR were used to detect MIR4435-2HG expression after transfection of si-YTHDC2 in SCC25 and CAL27 cells; (H) RT-qPCR were used to detect AC005562.1 expression after transfection of si-FTO in SCC25 and CAL27 cells; (I) RT-qPCR were used to detect the expression of COPDA1, LINC00271, MIAT after transfection of si-HNRNPC in SCC25 and CAL27 cells; (J) Kaplan–Meier plots showing the difference between OS in low and high-risk groups in training set; (K) the ROC curves and AUC values of the risk model for 1-, 3-, and 5-year survival in training set; (L) Kaplan–Meier plots showing the difference between OS in low and high-risk groups in testing set; (M) the ROC curves and AUC values of the risk model for 1-, 3-, and 5-year OS in testing set. (** p < 0.01, *** p < 0.001).