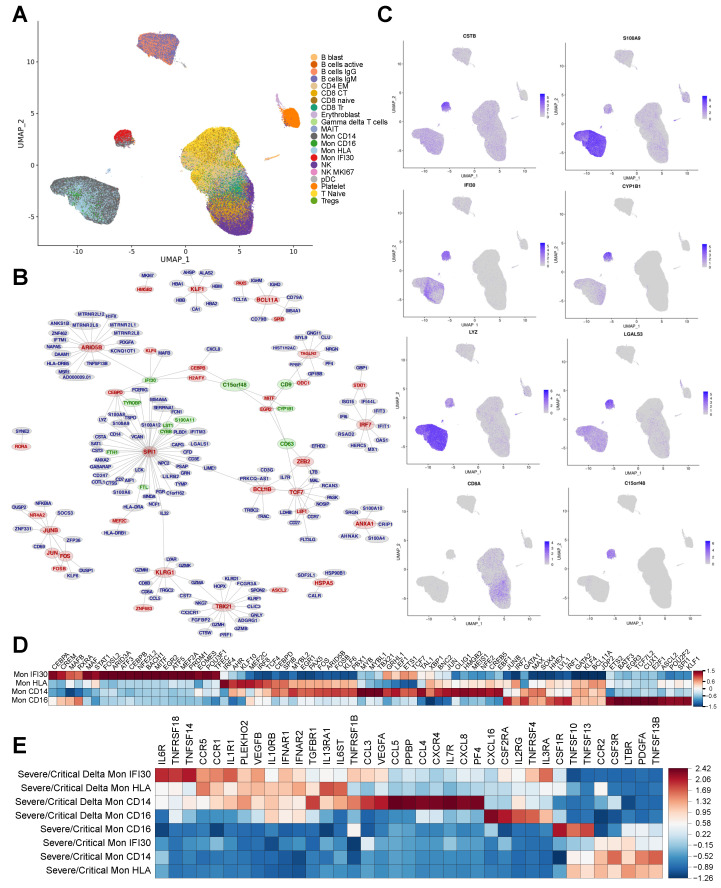
Figure 4.
(A) UMAP with colored cell annotation as obtained based on the CCA integration. Cell placing on the plot reflects similarities and colocalization of the cell types. (B) Fragment of the gene regulatory network reconstructed based on single–cell gene expression obtained for the whole dataset. Top 200 gene regulators (TFs) with their the most confident targets are plotted. Red color highlights important regulators that were recovered based on the SCENIC gene regulatory network reconstruction. Green labels show markers of the Mon IFI30 cell subtype. Network visualized using Fruchterman–Reingold layout algorithm. (C) Expression of the marker genes as projected on the SCENIC–based UMAP indicates activity of the core marker genes and colocalization of the cell subtypes. Mon IFI30 marker genes (LGALS3, CSTB, and C15orf48) are highly expressed in the population that was previously identified using CCA integration and clustering. (D) Heatmap with average gene expression for transcription factors that are differentially expressed (logFC > 0.75) in each cell type with respect to other monocytes. (E) Heatmap with average cytokine expression in monocytes for Delta and Wuhan-like samples.