Figure 1. Fungal DNA is present in multiple cancer types not explained by contamination, See also Figure S1.
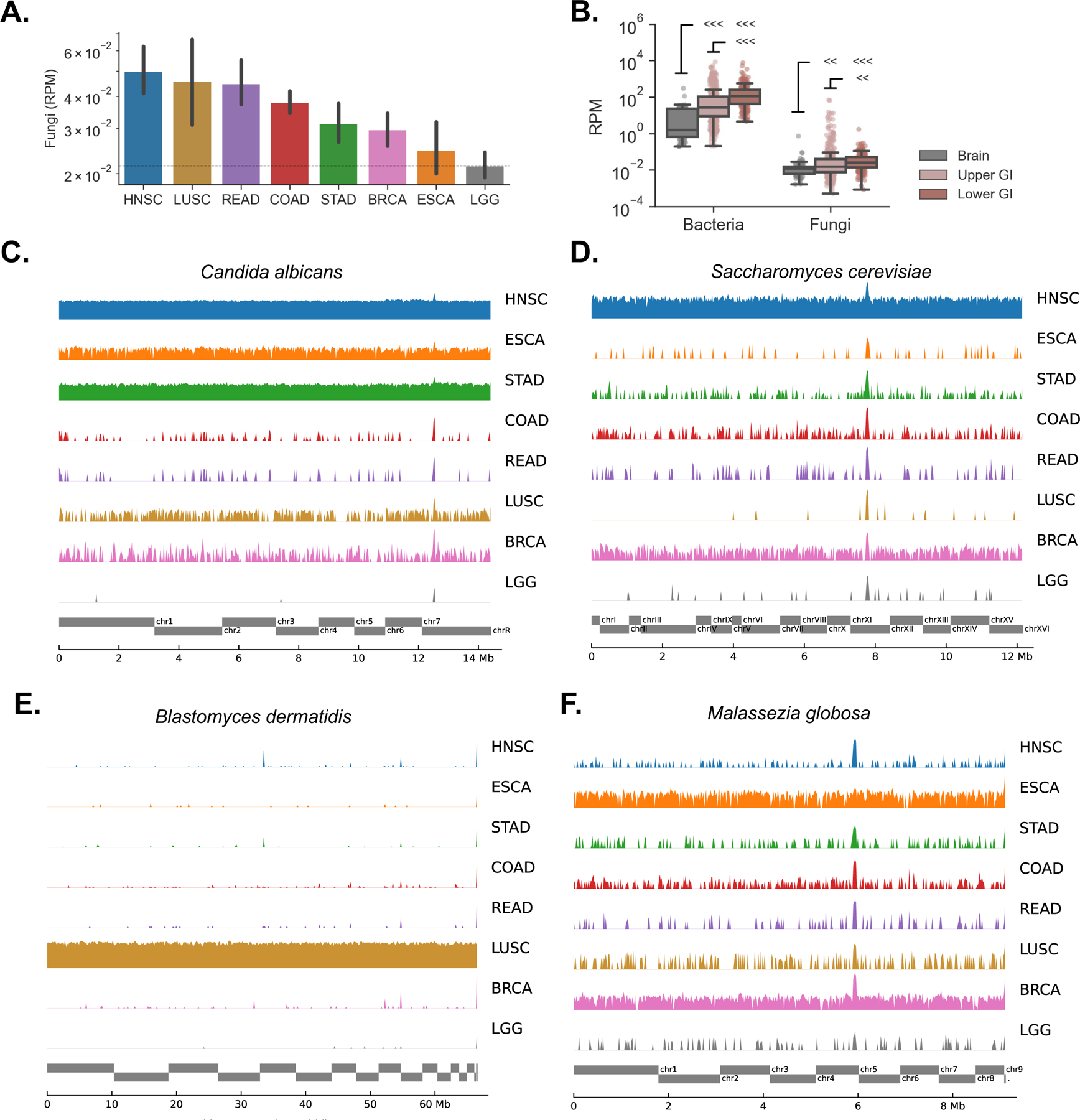
(A) Geometric mean of reads per million (RPM) of fungal DNA detected in tumor and tumor-associated tissue samples from head-neck (HNSC), lung (LUSC), rectum (READ), colon (COAD), stomach (STAD), breast (BRCA), esophageal (ESCA) and brain (LGG) cancers.
(B) Both bacterial and fungal reads were more abundant in the lower GI tract (COAD, READ) than the upper GI tract (HNSC, ESCA, STAD), and were more abundant in both GI groups compared to the brain (LGG) that was used here as a negative control.
(C – D)Genome alignments to C. albicans (C) and S. cerevisiae (D) are largely absent in brain but present at high rates across other tumor types, especially upper GI.
(E) Genome alignments to B. dermatidis are found at high rates in lung tumors, but not elsewhere.
(F) The distribution of sequencing reads aligning to M. globosa displays similar depth across sequencing projects including brain. Reads are distributed randomly, a signature of biological contamination.
