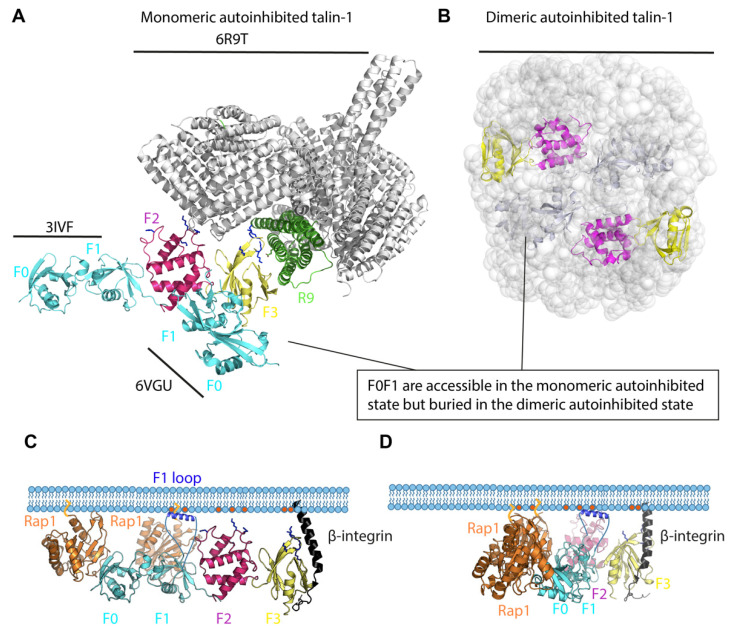
Figure 1.
Talin autoinhibition and activation. (A). Monomeric autoinhibited talin-1. The F0-F1 subdomains of the linear (PDB: 3IVF) or cloverleaf (PDB: 6VGU) talin FERM domain structures are superimposed onto the monomeric cryo-EM talin structure (PDB: 6R9T, grey). The subdomain interaction between the FERM domain and the rod domain, mainly F3 (yellow)–R9 (green), is critical for autoinhibition. F0-F1 subdomains, cyan; F2, magenta. (B). Dimeric autoinhibited talin-1. Talin-1 forms a compact autoinhibited conformation in which the talin rods form a donut-shaped structure and the two FERM domains (F0, F1, F2, F3) are packed in parallel and buried in the donut hole. F0-F1 subdomains, cyan; F2, magenta and F3, yellow. (C). Integrin–talin head–Rap1–plasma membrane complex. In the linear conformation shown here, the talin F0 and F1 (cyan) interact with two Rap1 molecules (orange), which are bound to the plasma membrane through their C-terminal geranyl-geranyl moieties. The F1 loop (dark blue) and positively charged patches in F2 and F3 interact with the negatively charged PIP2 phospholipids (red) in the membrane. (D). A model of integrin–talin head–Rap1–plasma membrane complex based on the cloverleaf talin-1 FERM domain. It is not known whether the linear and cloverleaf conformations exist exclusively, in parallel, or sequentially (see text for discussion).