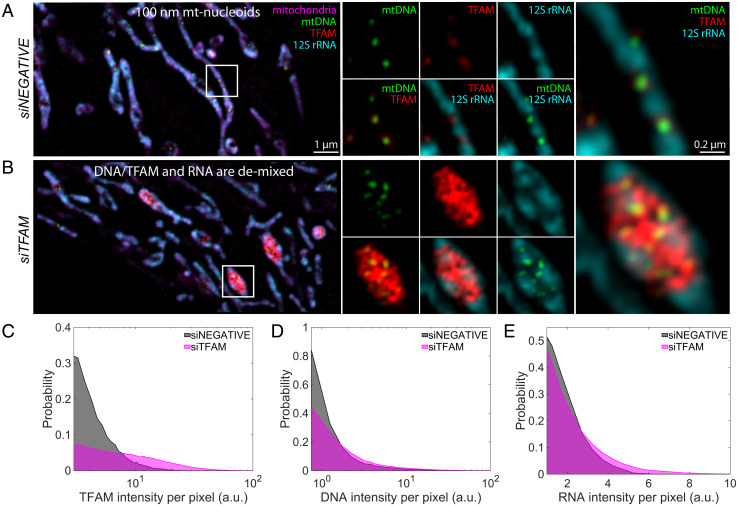
Fig. 4.
Depletion of mt-transcription components in vivo. (A and B) SIM images of mt components after 72 h of siRNA treatment: siNEGATIVE (A) and siTFAM (B). Left are the merged images of the zoomed-out version of the mitochondrial network, where mitochondria are in magenta (MitoTracker Red), mtDNA is in green (anti-DNA), TFAM is in red (anti-TFAM), and 12S rRNA is in cyan (RNA FISH). (Scale bar, 1 µm.) White box indicates region of interest (ROI). Middle are signal or two-channel overlays of the ROI. Right are the three-channel overlays for the ROI. (Scale bar, 0.2 µm.) Intensities across each channel are matched across siRNA conditions; n = 4 independent experimental replicates. (C) Probability distribution of TFAM pixel intensity within a segmented mt-nucleoid, where gray denotes siNEGATIVE, and magenta denotes siTFAM. (D) Probability distribution of DNA pixel intensity within a segmented nucleoid. (E) RNA pixel intensity surrounding mt-nucleoids. Data represent n = 4 independent experimental replicates that were pooled together.