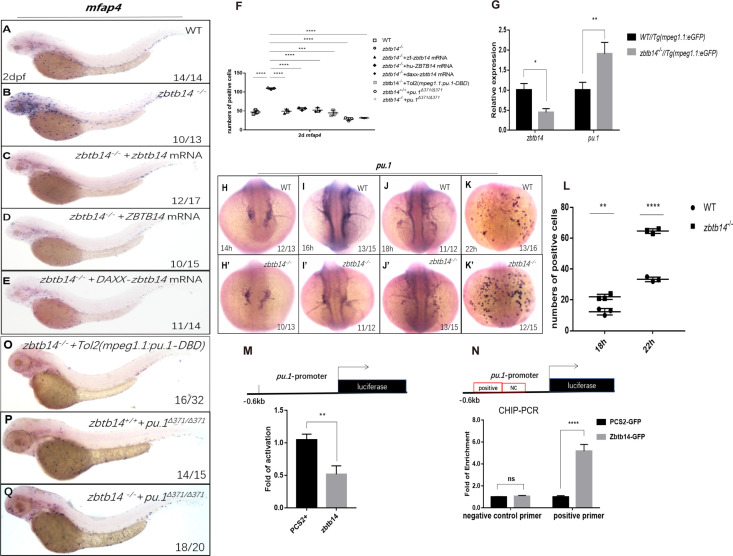
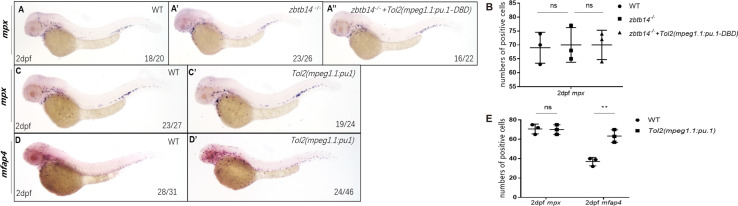
Figure 4. Zbtb14 regulates monocyte and macrophage development through inhibiting the expression of pu.1.
(A–E) mRNA rescue assays in zbtb14-/- larvae. mfap4 probe was used in whole-mount in situ hybridization (WISH) to examine rescue effect with wild zbtb14 (C), ZBTB14 (D), DAXX-ZBTB14 (E) mRNA injections. (F) Statistical result for A–E and O–Q. The statistical significance was calculated by using one-way analysis of variance (ANOVA). The statistical significance was calculated using one-way ANOVA followed by Dunnett T3 correction. The asterisk indicates a statistical difference. (N=3, 10–32 larvae were used for each experiment. Each dot represents the mean value of one experiment. Error bars represent mean ± standard error of the mean (SEM). ****p<0.0001.) (G) Quantitative reverse transcriptase polymerase chain reaction analysis of zbtb14 and pu.1 in GFP positive cells enriched from Tg(mpeg1.1:eGFP) and zbtb14-/-//Tg(mpx:eGFP) larvae at 2 days post-fertilization (dpf). To determine the relative expression rate, data were normalized to the expression level of wild type (WT) groups (which were set to 1.0) after normalized to the internal control of β-actin (Student’s t test, N=3. Error bars represent mean ± SEM. *p<0.05, **p<0.01). (H–K’) Serial WISH analyses of pu.1 in WT and zbtb14-/- embryos. (L) Statistical results for H–K’ (Student’s t test, N=3, 10–16 embryos were used. Each dot represents the mean value of one experiment, which was obtained from the counts of all of the embryos in the same group. Error bars represent mean ± SEM. **p<0.01, ****p<0.0001). (M) Luciferase reporter assay of Zbtb14 on the pu.1 promoter. Bars showed the relative luciferase activity on the zebrafish pu.1 promoter (–0.6 kb). (Student’s t test, N=3. Error bars represent mean ± SEM. **p<0.01.) (N) Chromatin immunoprecipitation polymerase chain reaction (ChIP-PCR) analysis of pu.1 promoter in zebrafish larvae expressing GFP or Zbtb14-GFP using an anti-GFP antibody. Positive: the location of the positive primers. NC: the location of the negative control primers. The statistical significance was calculated by using one-way ANOVA. The asterisk indicates a statistical difference. (N=3. Error bars represent mean ± SEM. ns: not statistically significant, ****p<0.0001.) (O–Q) WISH assay of mfap4 in zbtb14-/- mutants injected with TOL2 mpeg1.1:Pu.1 DBD, pu.1Δ371/Δ371 mutants, and zbtb14-/-//pu.1Δ371/Δ371 double mutants.
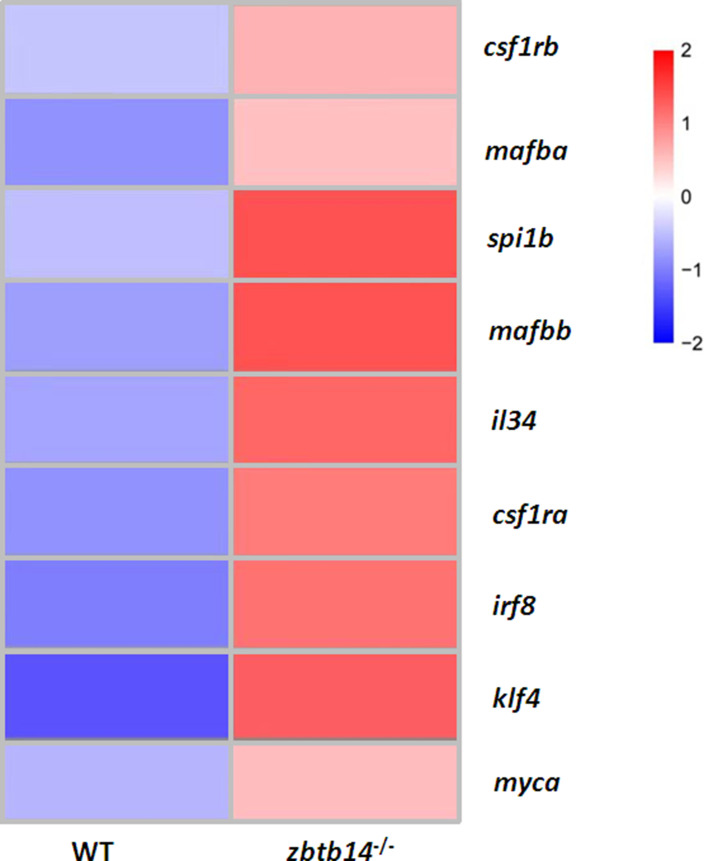
Figure 4—figure supplement 1. Heat map of genes associated with monocyte/macrophage development in mpeg1.1+ cells isolated from Tg(mpeg1.1:eGFP) and zbtb14-/-//Tg(mpeg1.1:eGFP) larvae at 2 days post-fertilization (dpf).
Figure 4—figure supplement 2. The role of zbtb14 in macrophage proliferation is lineage specific.
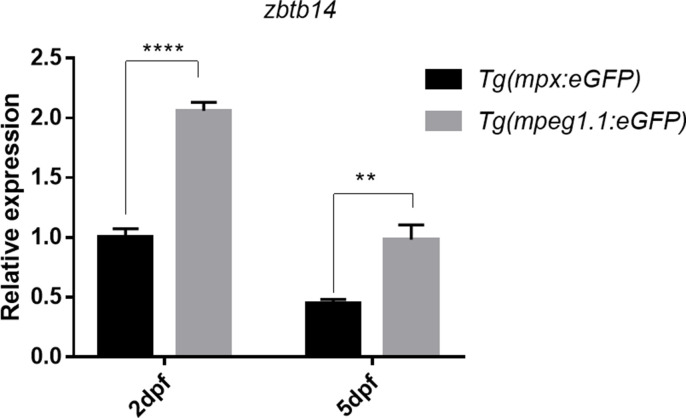
Figure 4—figure supplement 3. Expression of zbtb14 transcript in mpx+ and mpeg1.1+ cells at different developmental stages.
Figure 4—figure supplement 4. Luciferase reporter assay of Pu.1 on the zbtb14 promoter.
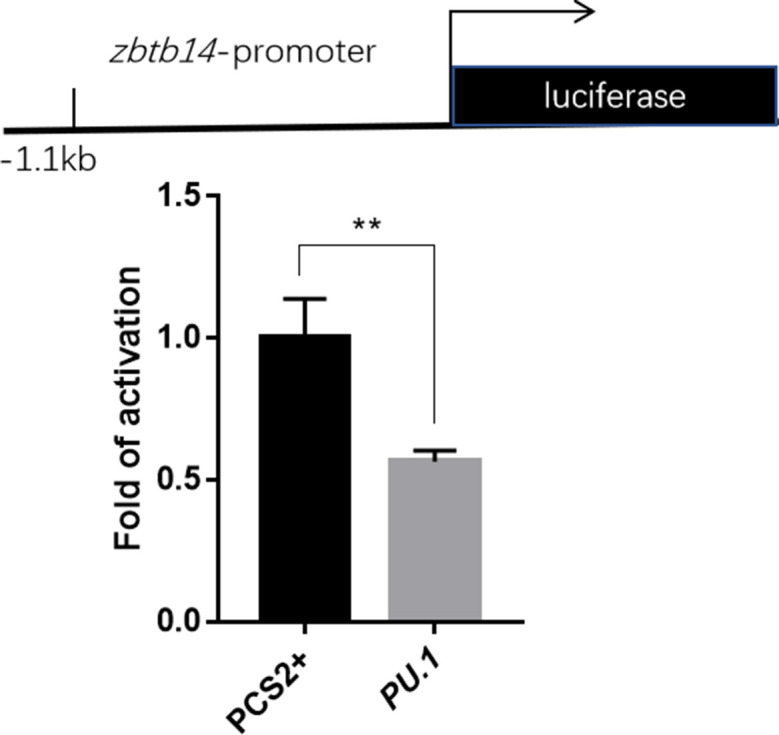
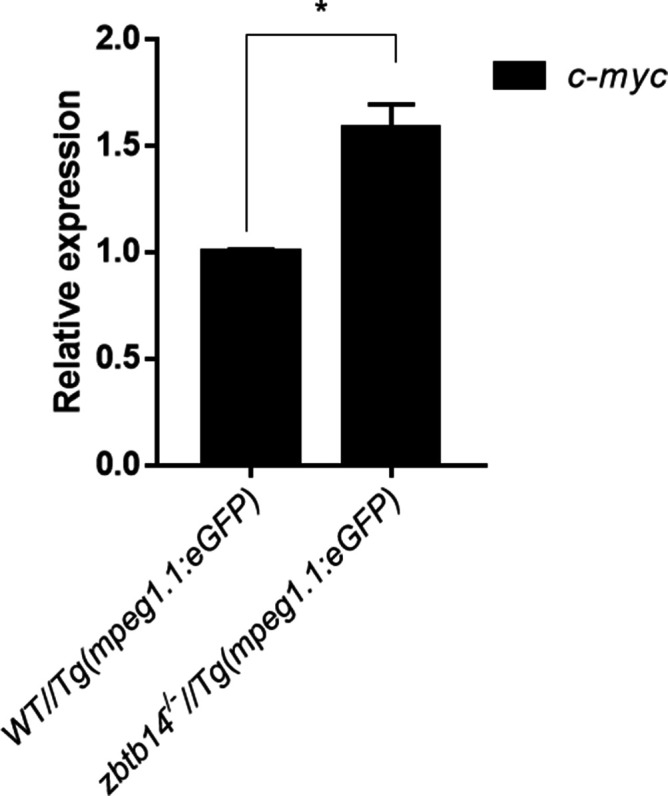
Figure 4—figure supplement 5. Expression of c-myc transcript in mpeg1.1+ cells.