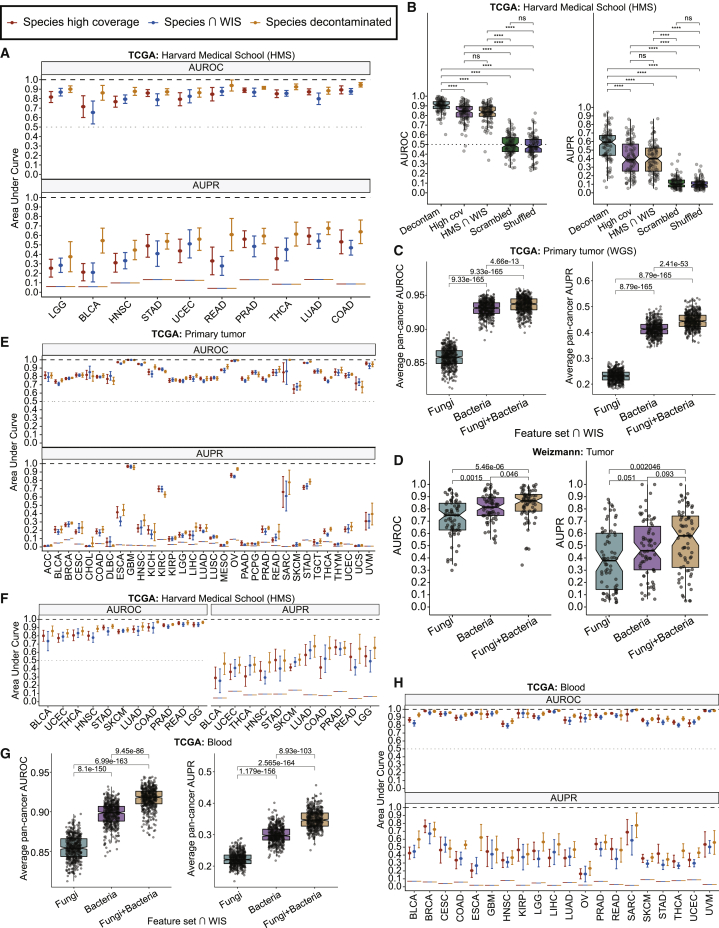
Figure 5.
Machine learning (ML) analyses reveal cancer-type-specific tumor and blood mycobiomes
(A) One-cancer-type-versus-all-others predictions on Harvard Medical School tumors (HMS, n = 876).
(B) Negative control analyses for (A) using scrambled metadata or shuffled samples. All one-cancer-type-versus-all-others performances are aggregated. ∗∗∗∗ q < 0.001; ns, not significant.
(C) Multi-class pan-cancer discrimination among TCGA WGS tumor samples using WIS-overlapping features across 500 independent folds (50 iterations of 10-fold CV).
(D) Aggregated one-cancer-type-versus-all-others ML performance in WIS cohort tumors.
(E) One-cancer-type-versus-all-others predictions using batch-corrected, TCGA primary tumor data (n = 10,998).
(F) One-cancer-type-versus-all-others predictions using HMS blood samples (n = 835).
(G) Multi-class pan-cancer discrimination among TCGA WGS blood samples using WIS-overlapping features across 500 independent folds (50 iterations of 10-fold CV).
(H) One-cancer-type-versus-all-others predictions using batch-corrected, TCGA blood data (n = 1,771).
(A, E, F, and H) Area under ROC curve (AUROC) and area under precision-recall curve (AUPR) measured on independent holdout folds (10-fold cross-validation [CV]) to estimate averages (dots) and 95% confidence intervals (brackets). “High coverage,” 31 fungal species with ≥1% aggregate genome coverage; “∩ Weizmann,” 34 WIS-overlapping fungal species; “decontaminated,” 224 decontaminated fungal species. Horizontal lines denote null AUROC or AUPR.
(B, C, D, and G) Two-sided Wilcoxon tests with Benjamini-Hochberg correction. Boxplots show median, 25th, and 75th percentiles and 1.5 × IQR.