Figure 1.
Zfp42R genes emerged with divergent expression in Fat1’s ancient TAD regulatory landscape
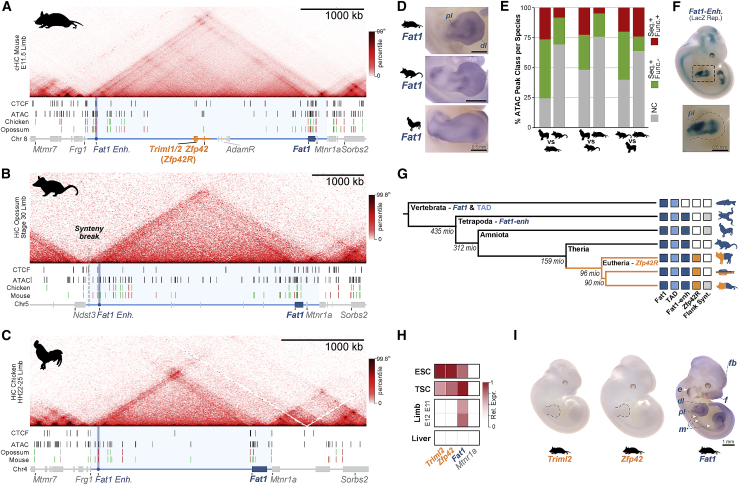
(A–C) cHi-C or Hi-C from mouse (A), opossum (B), and chicken (C) embryonic limb buds with ATAC-seq and CTCF ChIP-seq peaks below. Genes are colored bars and lines indicate the TAD (light blue), the 293 kb sub-Zfp42 region (Zfp42R, orange), and sub-Adam region (AdamR, gray). An ultra-conserved Fat1 enhancer (Fat1-enh, blue circle) is also highlighted. ATAC peaks are colored by sequence conservation (seq) with or without matching functional ATAC signal (func.). Red (seq+, func.+); green (seq+, func.−); gray (seq−,func.−).
(D) Species-specific Fat1 WISH in embryonic limbs. n = 2–4. Scale bar, 0.5 mm.
(E) Quantification of pairwise conservation of species ATAC-seq peaks.
(F) LacZ reporter assay of mouse Fat1-enh in E11.5 embryos. n = 4 embryos.
(G) Phylogenetic tree with presence of Fat1, the TAD, Fat1-enh, Zfp42R, or flanking synteny outside the TAD indicated.
(H and I) Gene activity overview from Fantom5 CAGE expression (H) and WISH (I). Fat1 WISH staining is seen in the ear (e), mammary glands (m), face (f), forebrain (fb), distal limb (dl), and proximal limb (pl). Trophoblast stem cells (TSCs). Scale bar, 1 mm.
See Figures S1 and S2 and Tables S1, S2, and S6.