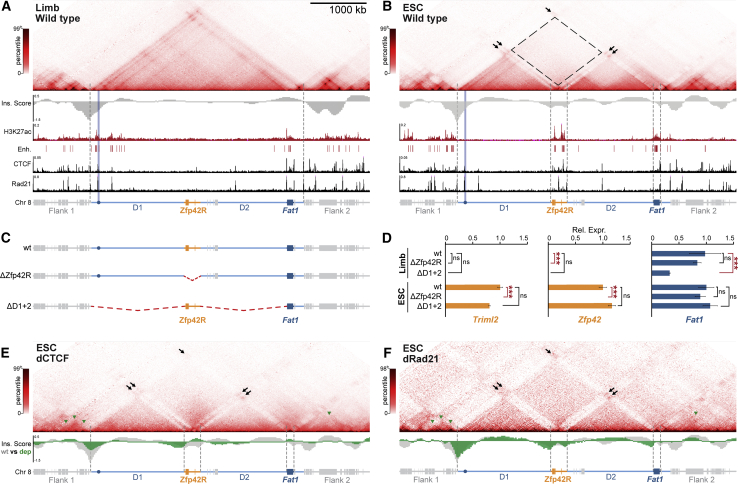
Figure 2.
Fat1 and Zfp42 independently utilize local enhancers in separated restructured domains in ESCs
(A and B) cHi-C from E11.5 limb buds (A) and ESCs (B) with insulation score (Ins. Score), H3K27ac, CTCF & Rad21 ChIP-seq, and called putative enhancers below. For cHi-C, black arrows indicate interactions between active H3K27ac-marked regions and dotted rectangle indicates lost interactions between inactive D1 and D2. E11.5 limb cHi-C is reproduced from Figure 1.
(C and D) Schematic of deletion mutants (C) with gene expression effects analyzed by RNA-seq (D). Error bars, SD calculated from 2–4 biological replicates per sample. ∗∗∗p < 0.001, ∗p < 0.05, non-significant (ns).
(E and F) cHi-C from dCTCF (E) or dRad21 (F) ESCs with wild type (gray) or depletion (green) Ins. Scores below. Green arrows indicate flanking TADs disrupted by CTCF/Rad21 depletion.