Figure 6.
Divergent promoter regulation is common in TADs throughout the genome
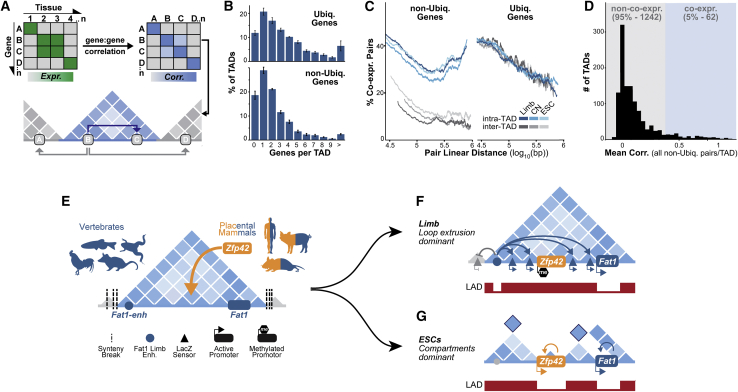
(A) Summary of TAD co-expression analysis. Gene pair co-expression was determined from FANTOM5 CAGE data, whereas TADs were identified in limb, cortical neuron (CN), and ESC Hi-C (Bonev et al., 2017; Consortium et al., 2014; Kraft et al., 2019; Lizio et al., 2015).
(B) Average frequency distribution of non-Ubiq. and Ubiq. genes in TADs.
(C) Fraction of co-expressing intra-TAD and inter-TAD gene pairs according to their linear separation. Lines represent a moving window average of 2,000 gene pairs.
(D) Frequency distribution of mean expression correlation between all non-Ubiq. genes in a domain for all multi-gene TADs.
(E–G) Model for evolution of independent Zfp42R and Fat1 regulation.
(E) Fat1, its enhancers, and TAD existed together as a regulatory unit in all vertebrates despite frequent flanking synteny breaks. Zfp42 and Triml1/2 emerged with independent regulation in placental mammals.
(F) In limbs, Fat1 enhancers emerge from LADs and promiscuously sample promoters throughout the domain's both active and NE-attached inactive compartments. However, despite this and its functional compatibility with Fat1 enhancers, DNA methylation of Zfp42’s promoter prevents its activation.
(G) In ESCs, activity-driven compartmentalization and perhaps weakened loop extrusion restructures the TAD, thereby driving the Zfp42R and Fat1 genes to independently utilize only local enhancers.
See Figure S6.