Figure S1.
Extended TAD evolutionary analysis and impact of Fat1-enh deletion, related to Figure 1
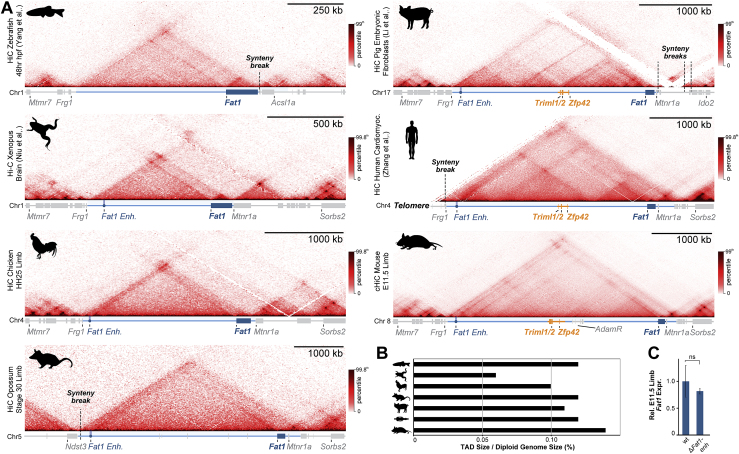
(A and B) Hi-C from species spanning the vertebrate family tree (A) with quantification of TAD:Diploid genome size (B) (Li et al., 2020; Niu et al., 2021; Yang et al., 2020; Zhang et al., 2019). Fat1 (dark blue box) has been universally maintained with a large gene desert and TAD (light blue line) whose size scales with diploid genome size. This is in spite of synteny breaks that relocate Mtnr1a (zebrafish), Frg1 (opossum), Mtmr7 (human), and Sorbs2 (pig). The limb Fat1-enh emerged in tetrapods while Mtnr1a and its isolated TAD became incorporated into Fat1’s TAD in the Mammalia lineage. Triml1, Triml2 and Zfp42 emerged in eutherian placental mammals where they are universally conserved within the ancient TAD. Finally, retroposition events created a cluster of disintegrin metalloproteinases (Adam26b, 26a and 34) within the Adam gene cluster specifically in rodents (Brachvogel et al., 2002; Choi et al., 2004; Long et al., 2012).
(C) RNA-seq expression effects of Fat1-enh deletion in E11.5 limbs. Error bars: standard deviation calculated from 4 biological replicates. non-significant (ns) p > 0.05.