Figure S3.
Confirmation of TAD disassembly in placental mammal pluripotency, CTCF/Rad21 depletion, and LAD signal strength, related to Figures 2 and 3
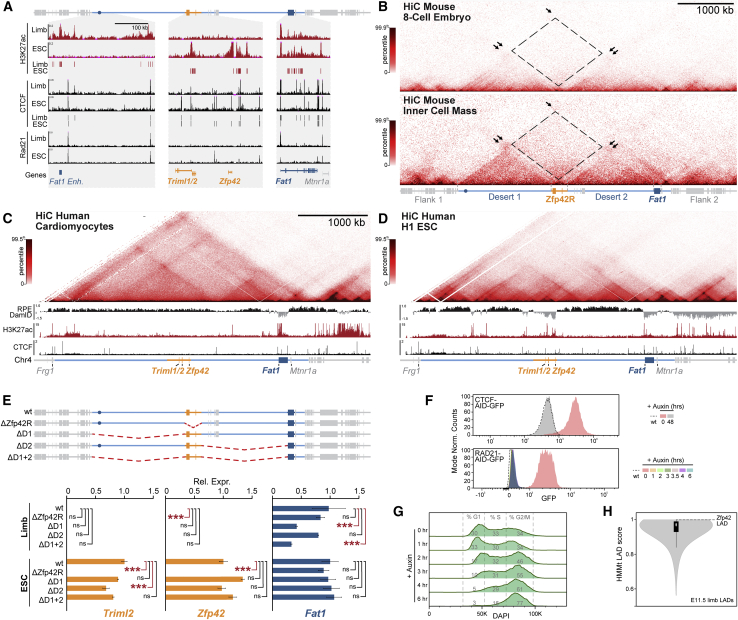
(A) Zooms of E11.5 limb and ESC H3K27ac, CTCF and RAD21 ChIP-seq with called enhancers or CTCF peaks below.
(B) Low input Hi-C from mouse 8-cell embryos (top) and pluripotent cells from the inner cell mass (bottom) (Du et al., 2017).
(C and D) Hi-C from human cardiomyocytes (C) and H1 ESCs (D) with corresponding H3K27ac, CTCF ChIP-seq and DamID shown below. Note DamID from retinal pigment epithelium (RPE) cells was used to define locus lamina-association when Zfp42R is inactive in differentiated cells.
(E) Schematic of deletion mutants (top) with effects on gene expression determined by RNA-seq (bottom). Error bars: standard deviation calculated from 2–4 biological replicates per sample. ∗∗∗p < 0.001, ∗p < 0.05, non-significant (ns).
(F) FACs distributions of GFP signal in CTCF-AID-GFP (top) and Rad21-AID-GFP (bottom) ESCs following indicated auxin treatments.
(G) Distribution of cell-cycle phases in Rad21-AID-GFP ESCs showing rapid accumulation in S and G2/M within 6 h. To account for accumulation of Rad21-AID-GFP ESCs in G2/M phase caused by failed sister chromatid cohesion, cHi-C was performed on sorted G1 cells 3.5 h post-auxin addition (Liu et al., 2021). By contrast, due to technical difficulties plating fixed cells on coverslips, FISH was performed on unsorted 2 h-induced Rad21-AID-GFP ESCs where only moderate shifts in the G1:S:G2/M ratio were observed.
(H) Genome-wide quantification of LAD scores from E11.5 limb DamID. The Zfp42 LAD is highlighted and lies in the 88th percentile of LADs genome wide.