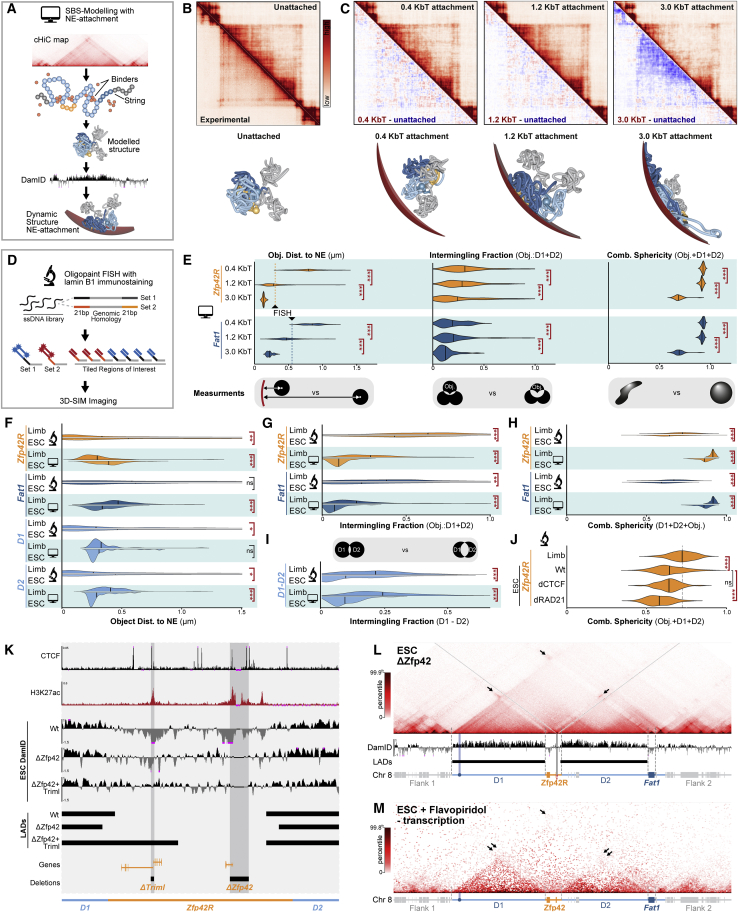
Figure S4.
Comparison of SBS modeling with NE attachment and Oligopaint FISH and summary of ESC H3K27ac-deletion mutants, related to Figure 3
(A) Schematic representation of the modified strings-and-binders (SBS) polymer model. cHi-C contact maps were used to define PRISM-assigned chromatin binders. The chromatin polymer is then structured in silco through simulated DNA interactions created by the self-association between matching binders (Barbieri et al., 2012; Nicodemi and Prisco, 2009). Generated structures were subsequently dynamically attached to a modeled NE with polymer affinities determined from sample-matched DamID (see STAR Methods).
(B and C) Reconstructed contact maps from simulated limb structures before (B) and after (C) NE attachment with 0.4, 1.2, and 3.0 kTb interaction energies. Corresponding subtraction maps and representative structures are shown below. n = 25–88 simulations.
(D) Oligopaint FISH 3D-SIM imaging strategy. A library of single stranded DNA oligos with genomic homology and overhangs allow multiplexed staining of multiple regions of interest.
(E) Quantification of object NE-distance (left), intermingling fraction (middle) and sphericity (right) for simulated limb structures following at indicated NE-attachment energies. 1.2 kTb was selected for further analysis as it produced NE-proximities without deforming the structure’s intermingling or sphericity relative to FISH measurements.
(F–H) Comparison of simulated NE-attachment model at 1.2 kTb and experimental FISH data in wild-type E11.5 limbs and ESCs. Measurements are object NE-distance (F), intermingling fraction (G), and object sphericity with D1+D2 (H).
(I) Comparison of simulated and observed D1 and D2 intermingling fraction.
(J) Quantification of combined FISH sphericity of Zfp42R with D1+D2 in indicated samples. Gray line highlights median limb values for reference. ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05 from Welch's t test comparisons. Non-significant (ns). FISH; n = 16–138 alleles of at least two biological replicates.
(K) Zooms of Zfp42R with indicated ESC H3K27ac, CTCF ChIP-seq, Lamin B1 DamID tracks below. Shaded boxes highlight deleted H3K27ac regions.
(L) cHi-C and DamID in ΔZfp42 ESCs.
(M) Published Micro-C of JM8.N4 ESCs where transcription is inhibited by flavopiridol (Hsieh et al., 2020). Arrows indicate Trim1/2, Zfp42 or Fat1 interactions with active chromatin and evasion of heterochromatin.