Figure S6.
Summaries of co-expression analysis and genome-wide effects of DNA hypomethylation, related to Figure 6
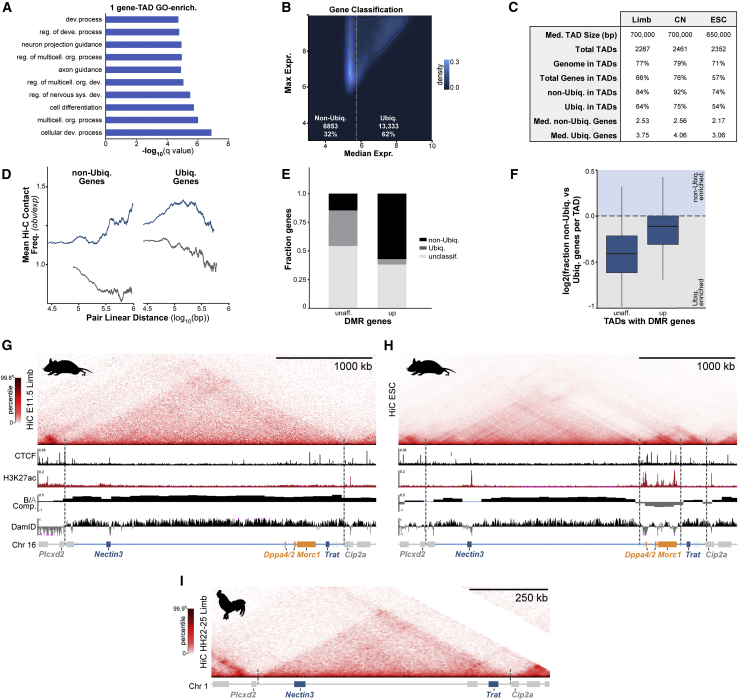
(A) GO-term enrichment for genes within single-gene TADs (Eden et al., 2009).
(B) Classification of genes into non-ubiquitously (non-Ubiq.) and ubiquitously (Ubiq.) expressed classes according to their maximum and median expression across FANTOM5 CAGE samples.
(C) TAD and gene statistics in limb, CNs and ESCs.
(D) Mean observed/expected KR-normalized Hi-C contact frequency between intra-TAD or inter-TAD gene pairs. Lines represent a moving window average of 2,000 gene pairs. Non-Ubiq. gene co-expression strongly correlates with their increased contact frequency within TADs and, in particular, near TAD boundaries.
(E) non-Ubiq. and Ubiq. expression classification of genes that possess hypomethylated DMR promoters in DNMT3B−/− limbs. Unclassified reflects genes that were detected in limb RNA-seq but did not pass thresholds for classification into Ubiq. or non-Ubiq. FANTOM5 classes.
(F) Fraction of non-Ubiq. versus Ubiq. genes in each TAD of hypomethylated DMR promoters.
(G–I) Hi-C at the Dppa2/4 locus from E11.5mouse limb buds (G), mouse ESCs (H) morphologically stage-matched chicken limb buds (I). Matching CTCF and H3K27ac ChIP-seq, compartments and Lamin B1 DamID tracks are shown below. Dotted lines demarcate partitioned domains. Nectin3 and Trat (dark blue) occupy a large gene desert and TAD (light blue) into which Morc1 (orange) emerged in tetrapods. Dppa2 and 4 (orange) emerged later in eutherians. Like Zfp42R genes, Dppa2/4 and Morc1 are active in ESCs where they are isolated with local enhancers in a separate domain within a disassembled TAD (Sima et al., 2019).